Use GenomeDK
An introduction to the GDK system and basic commands tinyurl.com/GDKslides
Bioinformatics Research Centre, Nat
GenomeDK, Health
Biomedicine, Health
2026-01-22
Some background
These slides are both a presentation and a small reference manual
We will try out some commands during the workshop
Official reference documentation: genome.au.dk
When you need to ask for help
Practical help:
Samuele (BiRC, MBG) - samuele@birc.au.dk
Drop in hours:
- Bioinformatics Cafe: https://abc.au.dk, abcafe@au.dk
- Samuele (BiRC, MBG) - samuele@birc.au.dk
General mail for assistance
support@genome.au.dk
Program
10:00-11:00: What is a HPC, GenomeDK, How it works, File System, virtual environments
11:00-12:00: desktop interface, new environment, transfer data, interactive job
12:45-14:00: Hands-on
Get the slides
Webpage: https://hds-sandbox.github.io/GDKworkshops/
Slides will always be up to date in this webpage
Navigate the slides
GenomeDK’s ABC
Learn your way around the basics of the GenomeDK cluster.
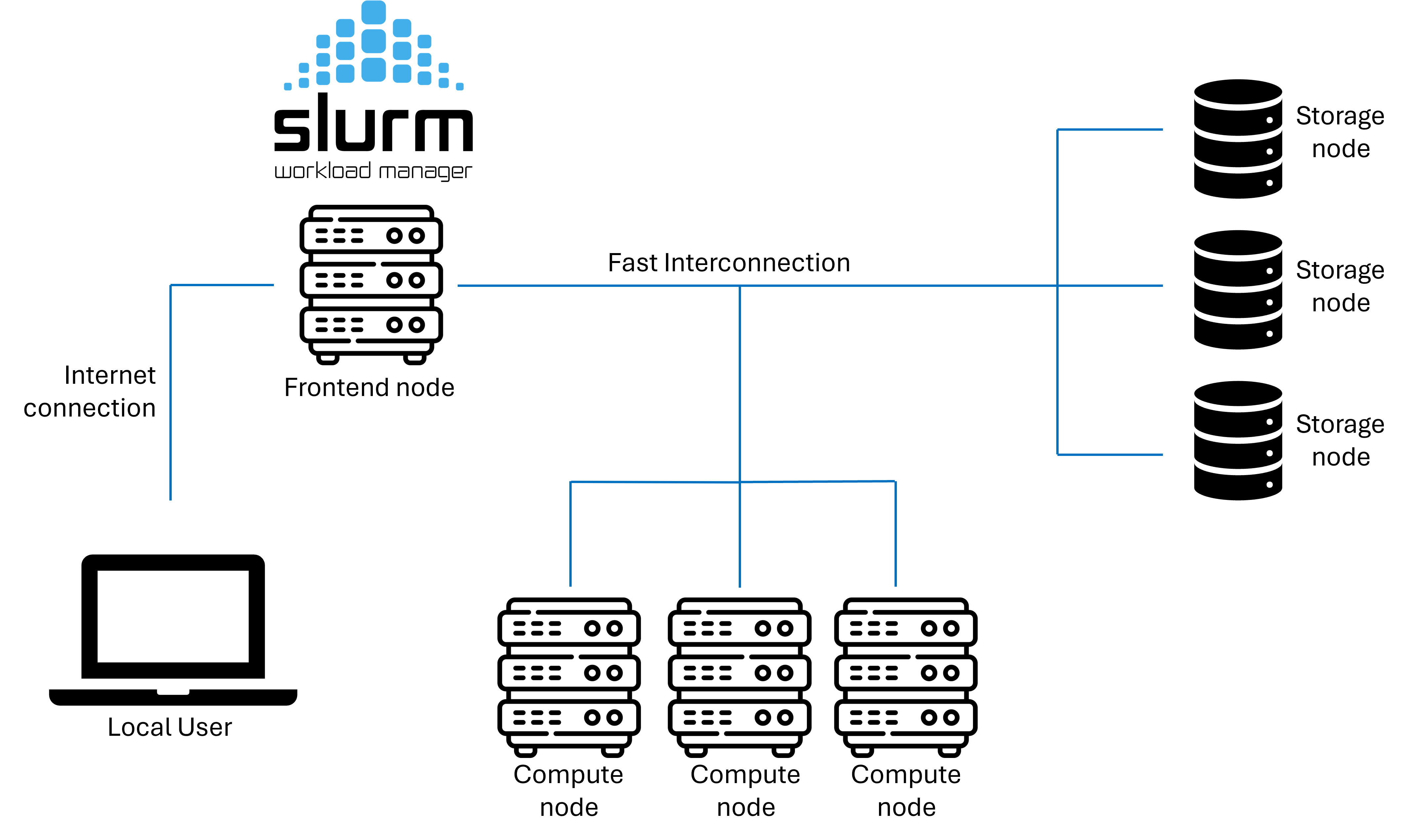
Infrastructure
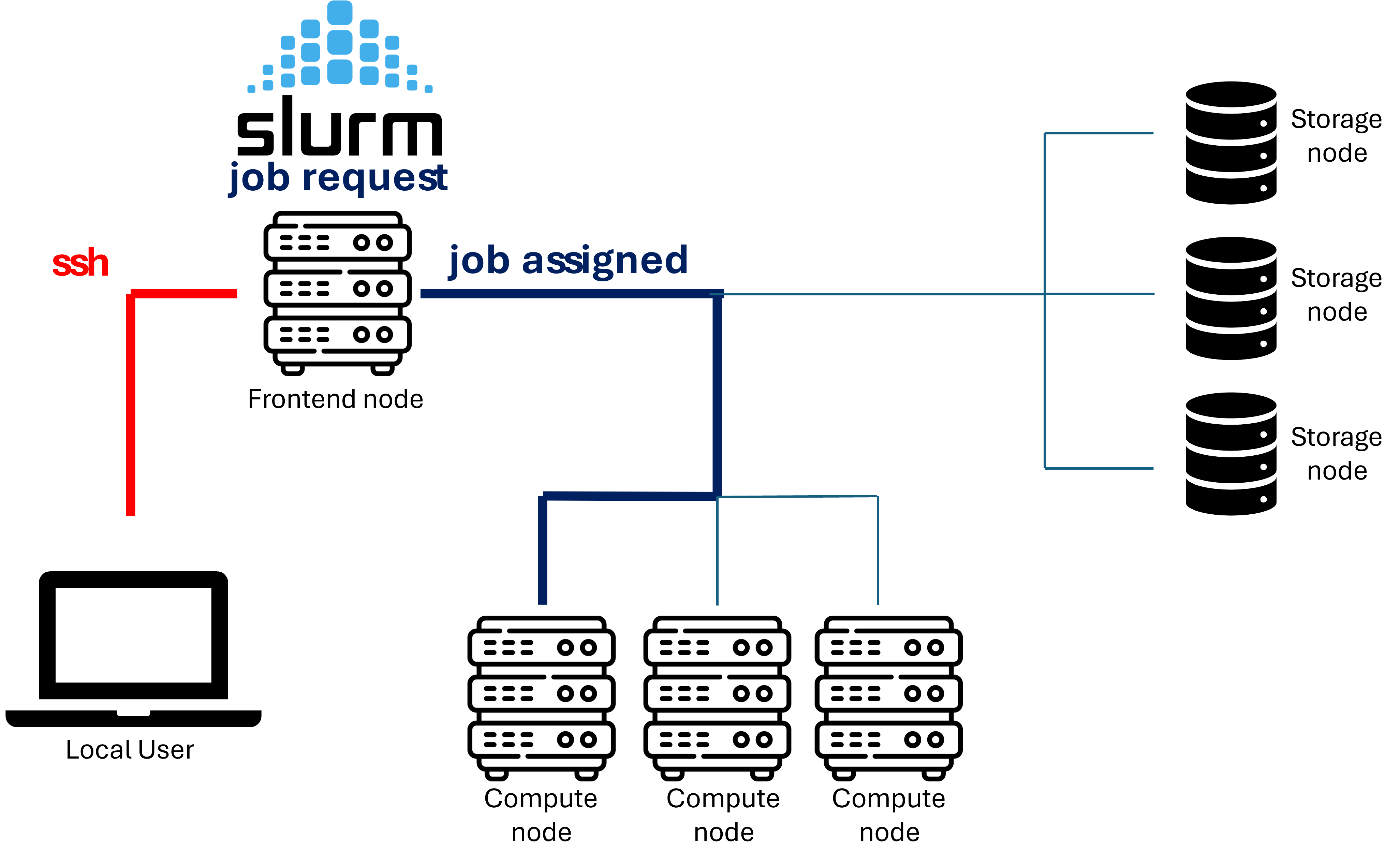
GenomeDK is a High performance computing (HPC) cluster, i.e. a set of interconnected computers (nodes). GenomeDK has:
- computing nodes used for running programs (~15000 cores, 16GPUs)
- storage nodes storing data in many hard drives (~23 PiB)
- a network making nodes talk to each other
- a frontend node from which you can send your programs to a node to be executed
- a queueing system called slurm to prioritize the users’ program to be run

HPC cluster from the backside

One node being mounted
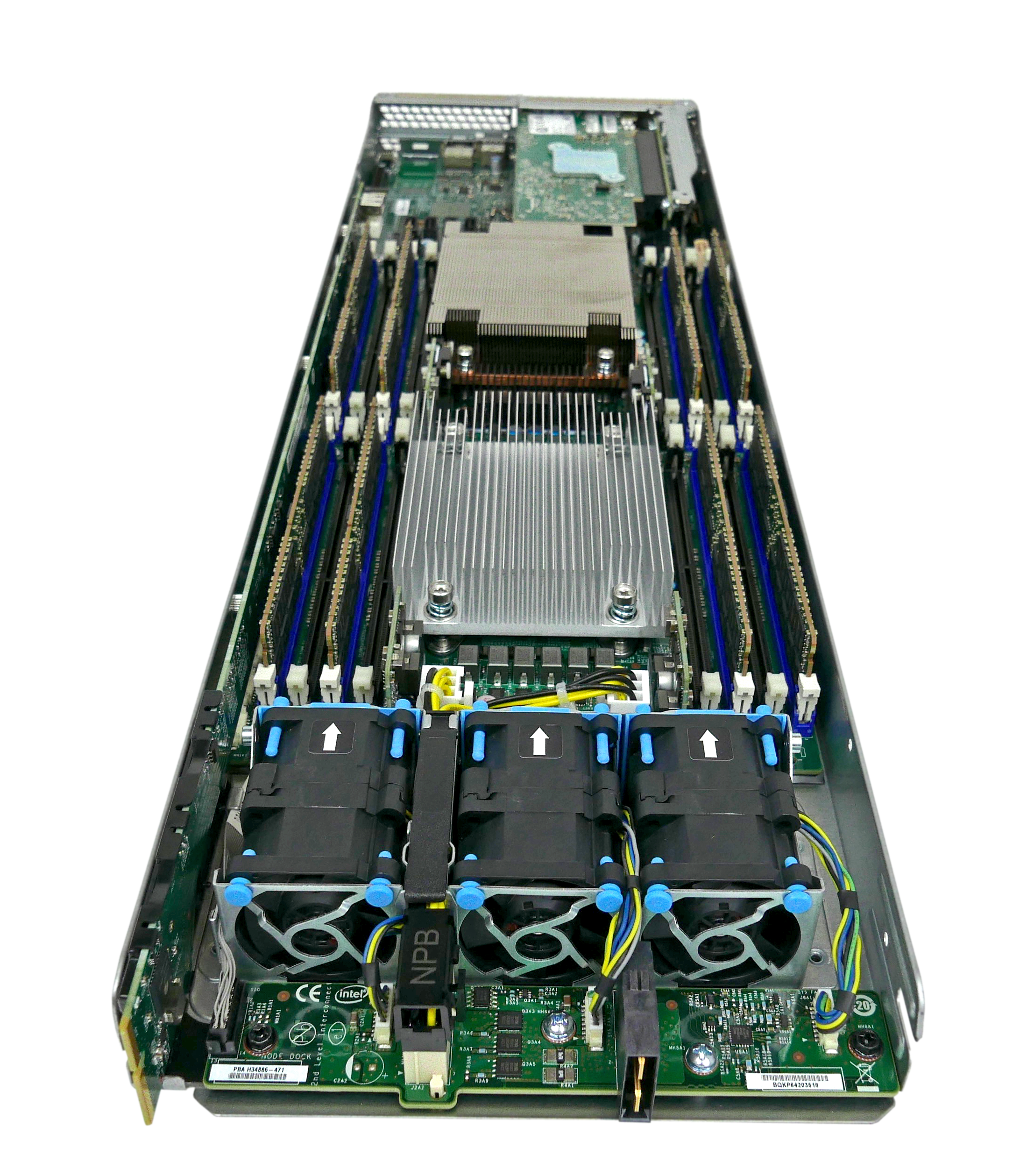
A single node with CPU and RAM
CPUs and cores
A CPU (Central Processing Unit) executes instructions from programs.
A CPU has multiple cores, each core can execute instructions independently at the cost of reduced bandwidth
Accessing a HPC
Powershell (Windows) or Terminal (MacOS, Linux) are powerful tools to interact with a HPC cluster.
Terminal for MacOS and Linux. This is called powershell in Windows.
Interactive desktop
GenomeDK also has an interactive desktop interface at desktop.genome.au.dk which can be used to access the cluster through a web browser. More on that later.
Terminals look old, but it gives you enormous flexibility through efficient commands and scripting possibilities.

An old “Dumb Terminal” from the 1970s
Getting access to GenomeDK
Creating an account happens through this form at genome.au.dk
![]()
Logging into GenomeDK happens through the command 1
When first logged in, setup the 2-factor authentication by
showing a QR-code with the command
scanning it with your phone’s Authenticator app 2.
Access without password
It is nice to avoid writing the password at every access. If you are on the cluster, exit from it to go back to your local computer
Now we set up a public-key authentication. We generate a key pair (public and private):
Always press Enter and do not insert any password when asked.
and create a folder on the cluster called .ssh to contain the public key
and finally send the public key to the cluster, into the file authorized_keys
After this, your local private key will be tested against GenomeDK’s public key every time you log in.
File System (FS) on GenomeDK
- Directory structure
- Absolute and Relative Paths
- important folders
- navigate the FS on the command line
How the FS is organized
Folders and files follow a hierarchy
/is the root folder of the filesystem - nothing is above that- the FS is shared across all machines and available to all users
homeandfaststorageare two of the folders in the root- projects are in
/faststorage/projectand linked to your home
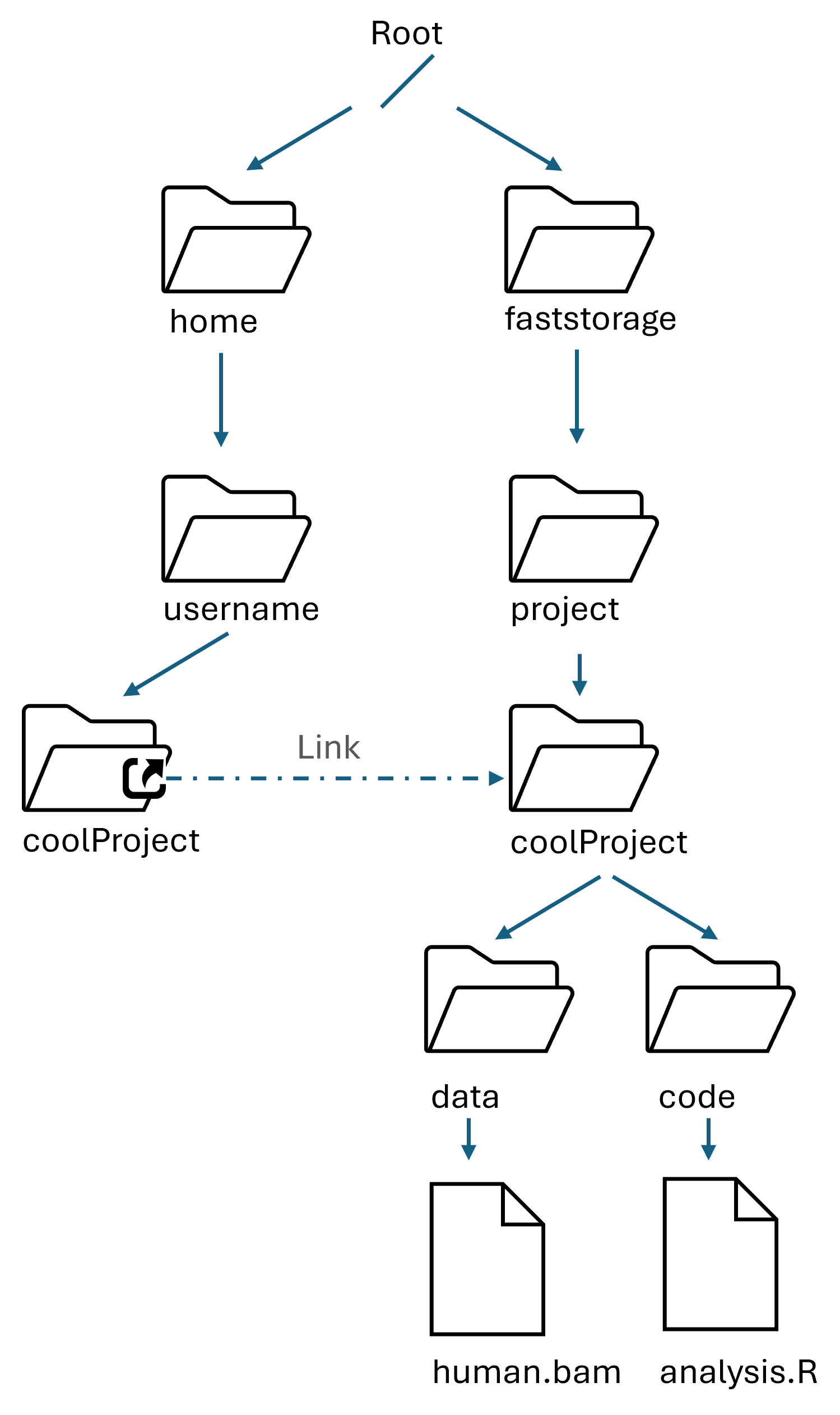
Exercise:
Log in: ssh USERNAME@login.genome.au.dk
Note
Run a command = Type a command + Enter
- Run
pwd, You should see your home folder:/home/USERNAME/home/USERNAMEis an example of path.pwdshows your current folder (WD, Working Directory)- you can write paths starting FROM the WD!
Run
ls .to show the content of your WD (the dot.)Run
mkdir -p GDKintroto create aGDKintrofolderRun
echo "hello" > ./GDKintro/file.txtto write hello in a fileUse
ls -lh ./GDKintroto see if the text file is there with some info.
Relative and absolute paths
/home/USERNAMEstarts from the root/. It is an absolute path../GDKintrostarts from the WD. It is a relative path.
Look at the File system tree and answer to the following questions:

Something more about your home
After log in, you will find yourself into your private home folder, denoted by ~ or equivalently /home/username. Your prompt will look like this:
which follows the format [username@node current_folder].
Warning
- Do not fill up your home folder with data. It has a limited amount of storage (a quota of 100GB).
- Your home folder is private to you only
Exercise cont’d
We now set the WD into GDKintro and remove all text files in it. Then we download a zipped fastq file, unzip it, and print a preview!
Some notes about the commands
rm *.txtremoves all files ending with.txt. The symbol*is a wildcard for the file nameForever away
There is no trash bin - removed files are lost forever - with no exception
headprints the first lines of a text file
Exercise: Read files
Useful utility 1: less file reader. less is perfect for exploring (big) text files: you can scroll with the arrows, and quit pressing q. Try
The very first sequence you see should be
@HISEQ_HU01:89:H7YRLADXX:1:1101:1116:2123 1:N:0:ATCACG
TCTGTGTAAATTACCCAGCCTCACGTATTCCTTTAGAGCAATGCAAAACAGACTAGACAAAAGGCTTTTAAAAGTCTA
ATCTGAGATTCCTGACCAAATGT
+
CCCFFFFFHHHHHJJJJJJJJJJJJHIJJJJJJJJJIJJJJJJJJJJJJJJJJJJJHIJGHJIJJIJJJJJHHHHHHH
FFFFFFFEDDEEEEDDDDDDDDDChallenge yourself
Search online man less (or with less --help) how to look for a specific word in a file with less. Then visualize the data with less, and try to find if there is any sequence of ten adjacent Ns (which is, ten missing nucleotides). Then, answer the question below
Exercise: Write files
Useful utility 2: nano text editor. It open, edits and saves text files. Very useful for changes on the fly.
Try
nano data.fastq. Change a base in the first sequence,then press Ctrl+O to save (give it a new file name
newData.fastqand press Enter)press Ctrl+X to exit. If you use
lsyou can see the new saved file.
Project management
- What are GDK projects
- how to track the resource usage, and
- how to organize a project
GDK projects
what is a project
Projects are contained in /faststorage/project/ and linked in your home, and are simple folders with some perks:
- you have to request their creation to GDK administrators
- access is limited to you, and users you invite
- CPU, GPU, storage and backup usage are registered under the project for each user
- you can keep track of per-project and -user resource usage
Common-sense in project creation
- Do not request a lot of different small project, but make larger/comprehensive ones
- No-go example: 3 projects
bulkRNA_mouse,bulkRNA_human,bulkRNA_apeswith the same invited users - Good example: one project
bulkRNA_studieswith subfoldersbulkRNA_mouse,bulkRNA_human,bulkRNA_apes.
- No-go example: 3 projects
- Why? Projects cannot be deleted, so they keep cumulating
Creation
Request a project (after login on GDK) with the command
After GDK approval, a project folder with the desired name appears in ~ and /faststorage/project. You should be able to set the WD into that folder:
or
Users management
Only the creator (owner) can see the project folder. You (and only you) can add an user
or remove it
Users can also be promoted to have administrative rights in the project
or demoted from those rights
Accounting
You can see globally monthly used resources of your projects with
Example output:
More detailed usage: by users on a selected project
You can see how many resources your projects are using with
Example output:
project period billing hours storage (TB) backup (TB) storage files backup files
ngssummer2024 sarasj 2024-7 77.98 0.02 0.00 528 0
ngssummer2024 sarasj 2024-8 0.00 0.02 0.00 528 0
ngssummer2024 savvasc 2024-7 223.21 0.02 0.00 564 0
ngssummer2024 savvasc 2024-8 0.00 0.02 0.00 564 0
ngssummer2024 simonnn 2024-7 173.29 0.01 0.00 579 0
ngssummer2024 simonnn 2024-8 0.00 0.01 0.00 579 0Accounting Tips
- You can pipe the accounting output into
grepto isolate specific users and/or months:
- all the accounting outputs can be saved into a file, which you can later open for example as Excel sheet.
Example:
Private files or folders
Folders management
Have a coherent folder structure - your future self sends manu thanks.
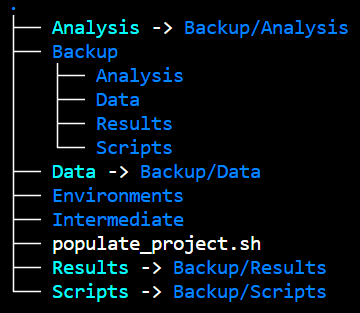
Example of structure, which backs up raw data and analysis
If your project has many users, a good structure can be

MUST-KNOWs for a GDK project
- remove unused intermediate files
- unused and forgotten object filling up storage
- backup only the established truth of your analysis
- in other words the very initial data of your analysis, and the scripts
- outputs of many files should be removed or zipped together into one
- otherwise GDK indexes all of them: slow!!!
Backup cost >>> Storage cost >> Computation cost
Software management
No preinstalled software on GenomeDK
You install and manage your software and its dependencies inside virtual environments
Virtual environments
Each project needs specific software versions dependent on each other for reproducibility - without interferring with other projects.
Definition
A virtual environment keeps project-specific softwares and their dependencies separated
A package manager is a software that can retrieve, download, install, upgrade packages easily and reliably
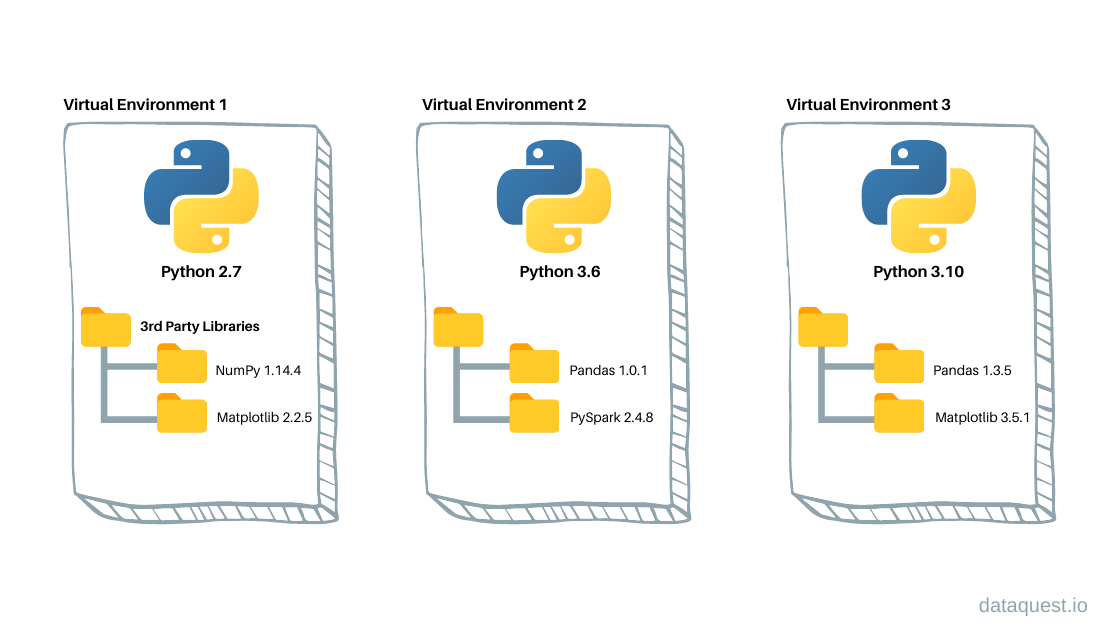
How virtual envs work: packages at different versions are kept separated into folders, together with all system files needed to make them work.
Conda
Conda is both a virtual environment and a package manager.
- easy to use and understand
- can handle quite big environments
- environments are easily shareable
- a large archive (Anaconda) of packages
- active community of people archiving their packages on Anaconda
Pixi
A newer virtual env and package manager
- An upgrade of Conda in speed and stability
- Can install the same packages as conda
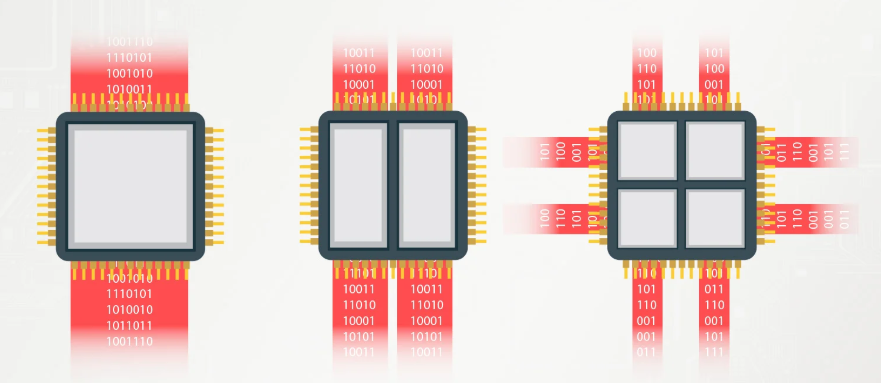
A package manager puts together the dependency trees of requested packages to find all compatible dependencies versions.
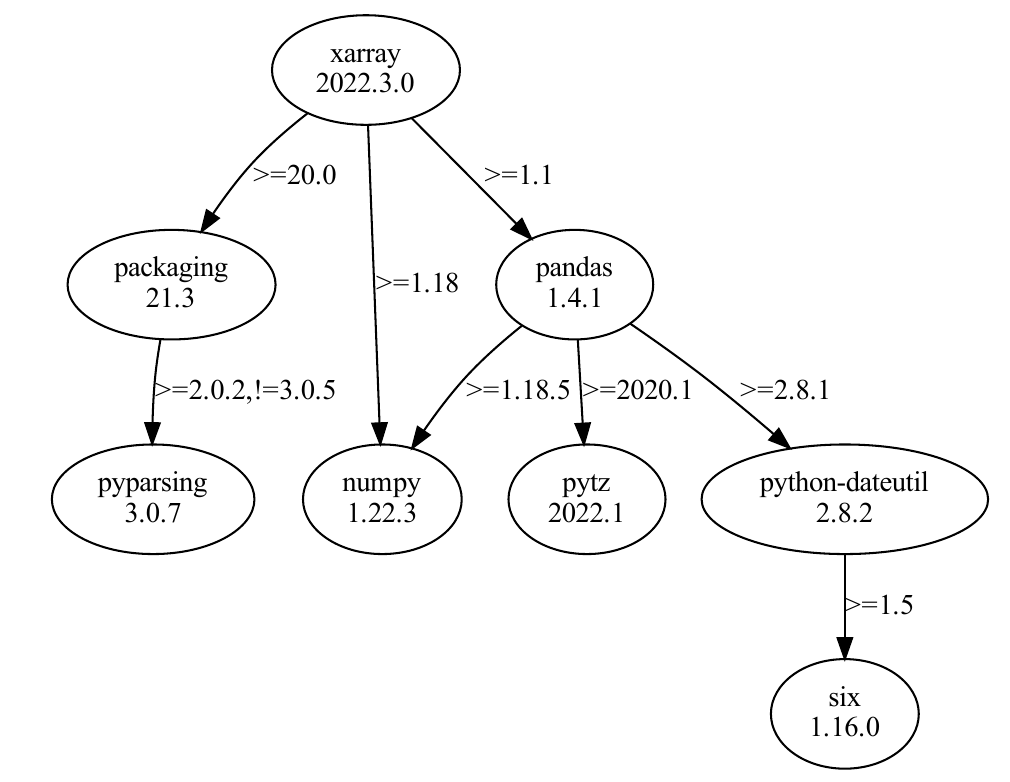
Figure: A package’s dependency tree with required versions on the edges
To install a specific package in your environment, search it on anaconda.org:
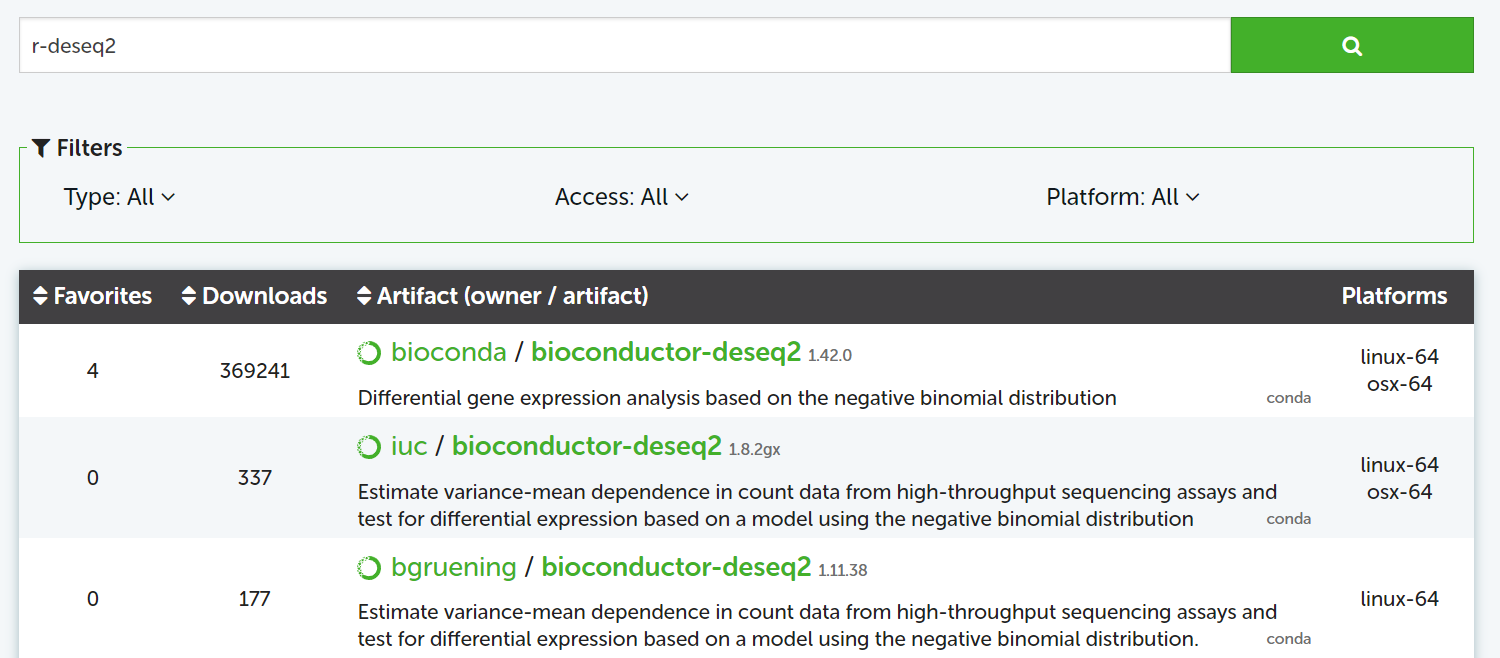
Figure: search DeSeq2 for R
Channels
packages are archived in channels. conda-forge and bioconda include most of the packages for bioinformatics and data science.
conda-forge packages are often the most up-to-date.
Exercise - Pixi
First of all, we open the desktop interface to GenomeDK at desktop.genome.au.dk. Choose the open frontend for the login.
The desktop session will be operative even if you close and reopen your browser afterwards!
The terminal will work as if you logged into the frontend (The desktop is logged into the front-end node already). You can also use the browser!
Open the terminal and run the command below to install pixi:
After that, make the system recognize pixi
Change your WD with the one we created earlier, where we have the file data.fastq
Initiate a new pixi environment into the folder:
This will also use conda-forge and bioconda as channels for package installation.
What are channels?
Channels are repositories where packages are stored.
conda-forge and bioconda are two of the most used channels for bioinformatics and data science and contain virtually any package you may need.
Note that we specified channels in the order of priority: conda-forge has higher priority than bioconda when installing packages, so you will always search for a package first in conda-forge and then in bioconda.
Use the file browser and open the GDKintro folder
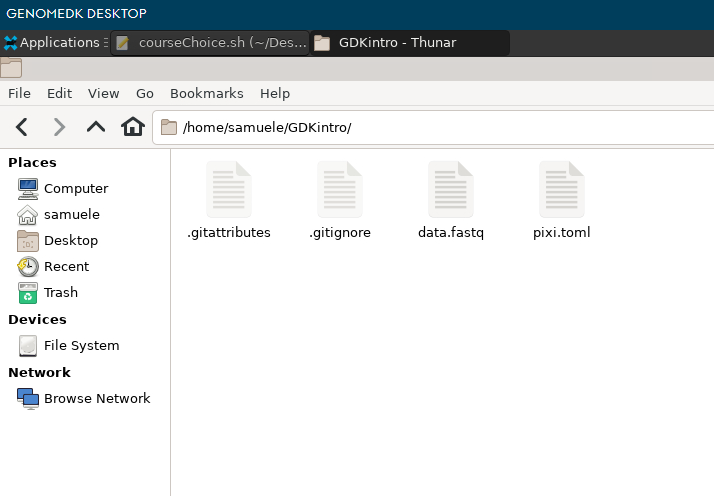
You can see some new files. pixi.toml contains info pixi will use to create your environment.
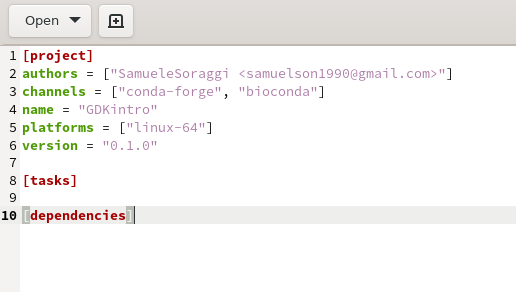
Open pixi.toml with the text editor, and make sure you have the two channels conda-forge and bioconda as you required with the pixi init command. You can always add more channels if you find out later that you need them for specific packages
Now get back to the terminal and install some packages. Your working directory MUST be the same where pixi.toml is located!
The terminal will look like this at the end
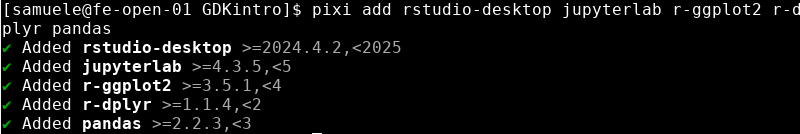
Now open the pixi.toml file. You should see all the installed packages with related information.
Exercise Cont’d
Be sure your WD is in the folder GDKintro. Then run
Open the file environment.yml. It looks very similar to pixi.toml and is compatible with conda to recreate your environment.
Let’s zip those files into one:
Data transfer
Data can be downloaded/uploaded in two ways:
from the command line of a local computer
using an interactive interface (Filezilla)
Exercise cont’d - Download on shell
How to download the environment files to our computer? Open a terminal on your computer and run this command:
scp needs your login and the absolute path to the file. We give also the download destination as the WD on the local computer (.)
Exercise - Download interactively
You can transfer data with an interactive software, such as Filezilla, which has an easy interface. Download Filezilla.
When done, open Filezilla and use the following information on the login bar:
- Host:
login.genome.au.dk - Username, Password: your
GenomeDKusername and password - Port:
22
Press on Quick Connect. As a result, you will establish a secure connection to GenomeDK. On the left-side browser you can see your local folders and files. On the right-side, the folders and files on GenomeDK starting from your home.
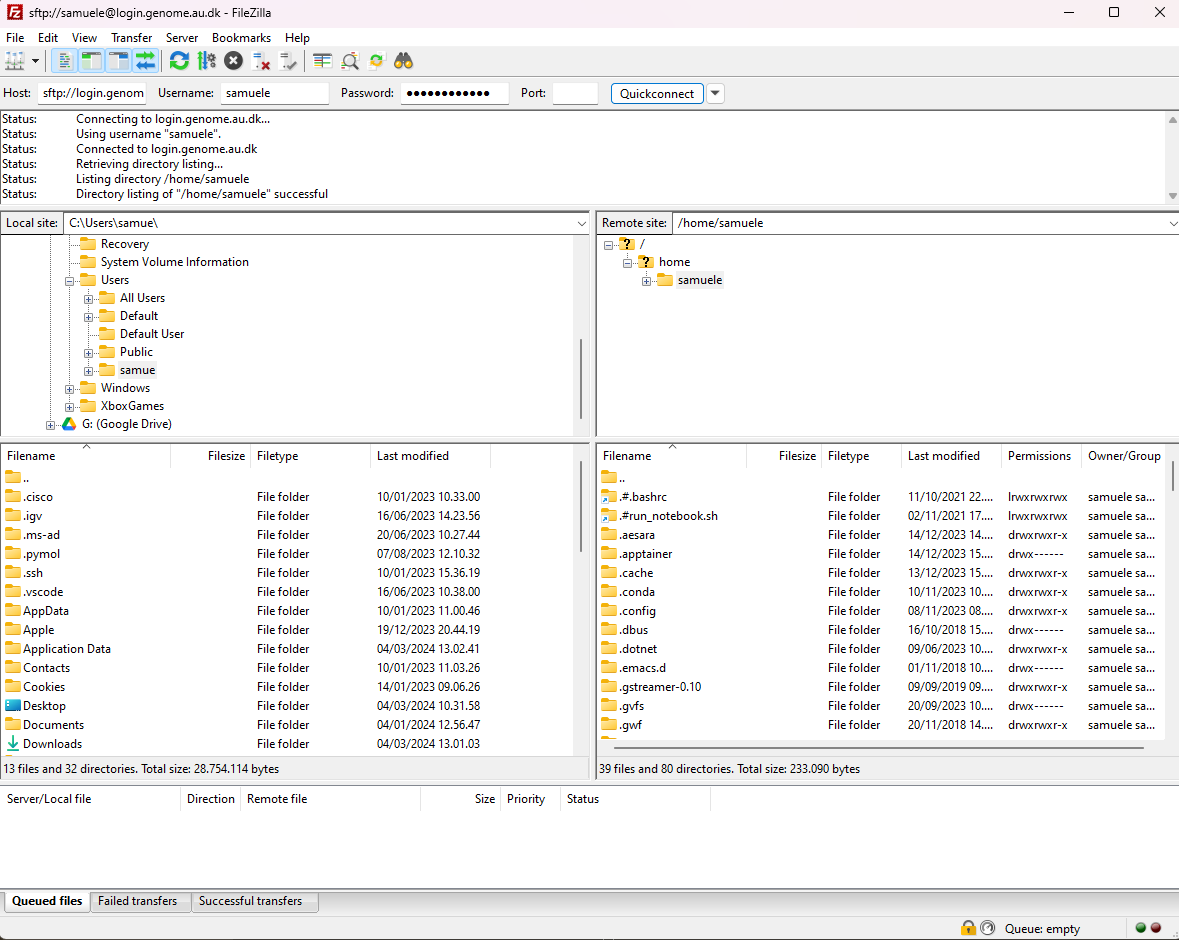
Download the environment.zip file. You need to right-click on it and choose Download
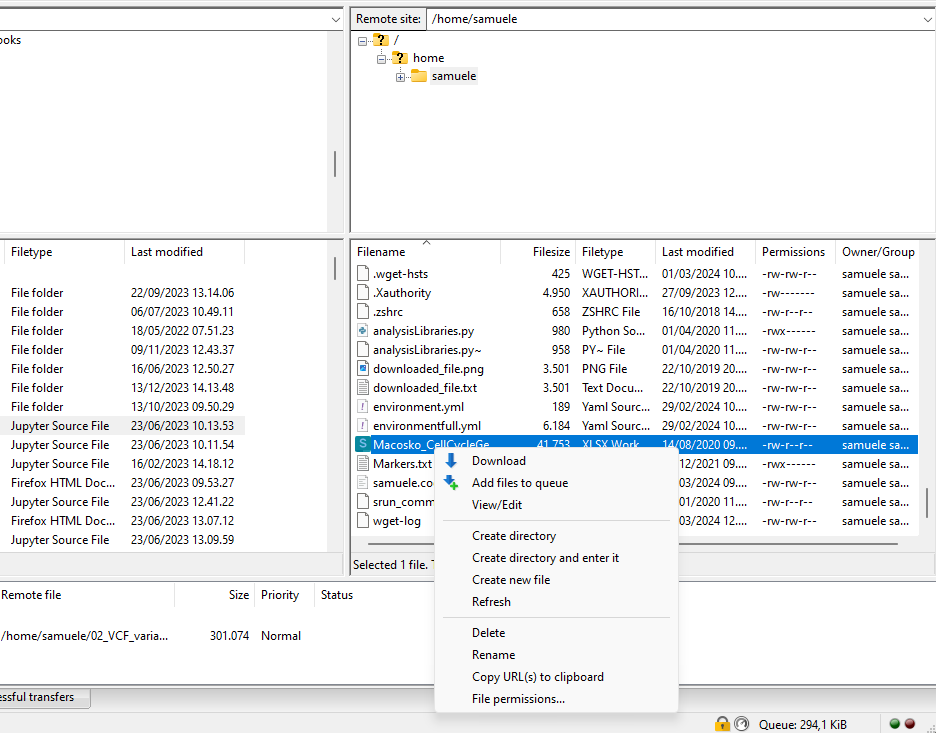
You can do exactly the same to upload files from your local computer!
Running a Job
Running programs on a computing cluster happens through jobs.
Learn how to get hold of computing resources to run your programs.
What is a job on a HPC
A computational task executed on requested HPC resources (computing nodes), which are handled by the queueing system (SLURM).
The command gnodes will tell you if there is heavy usage across the computing nodes
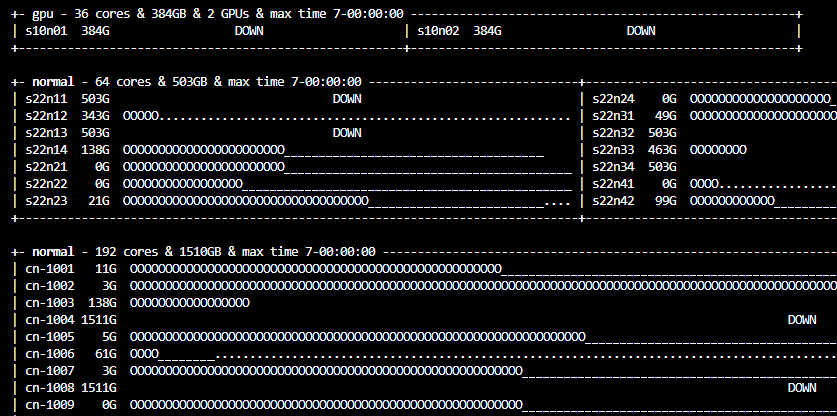
Usage of computing nodes. Each node has a name (e.g. cn-1001). The symbols for each node mean running a program (0), assigned to an user (_) and available (.)
If you want to venture more into checking the queueing status, Moi has done a great interactive script in R Shiny for that.
Front-end nodes are limited in memory and power, and should only be for basic operations such as
starting a new project
small folders and files management
small software installations
data transfer
and in general you should not use them to run computations. This might slow down all other users on the front-end.
Interactive jobs
Useful to run a non-repetitive task interactively
Examples:
splitting by chromosome that one
bamfile you just gotopen Rstudio and Jupyterlab
compress/decompress multiple files, maybe in parallel
Once you exit from the job, anything running into it will stop.
Exercise: Interactive job
You can also run an interactive job on GenomeDK desktop. Go back to it and use the terminal to go into the GDKintro folder:
Now run an interactive job. Use 8g of RAM, 2 cores, and choose 01:00:00 hours. Choose the account using the name of one of your projects, or delete it if you do not have projects.
You will have to wait in queue. When you get the resources, the node in use is shown in the prompt. Below, for example, the node is s21n32.
[USERNAME@s21n32 ~]$Now, run rstudio or jupyterlab (your choice!) from the pixi environment:
The packages available in Rstudio and Jupyterlab are the ones installed in your environment. More on this will be in our Advanced GenomeDK workshop.
Closing the workshop
Please fill out this form :)
A lot of things we could not cover
use the official documentation!
ask for help, use drop in hours
try out stuff and google yourself out of small problems
Slides updated over time, use as a reference
Future workshops about advanced usage and pipelines