nf-core/rnaseq 3.17.0
💬 Learning Objectives:
- Learn about the UCloud computing system.
- Learn how to submit a job and explore your results folders.
- Submit a nf-core/rnaseq 3.17.0 run on our data
We will be running the nf-core/rnaseq pipeline. Please refer to their detailed documentation here: https://nf-co.re/rnaseq/3.17.0/. We highly recommend reading through all the sections to fully understand how to run the pipeline, explore the optional parameters you can set, and learn about the output format.
Submit a job
Access Ucloud with your account and choose the project Sandbox RNASeq Workshop to which you have been invited (contact the team if you haven’t).
Click on Apps on the left-side menu, search for the application nf-core rnaseq, and click on it.
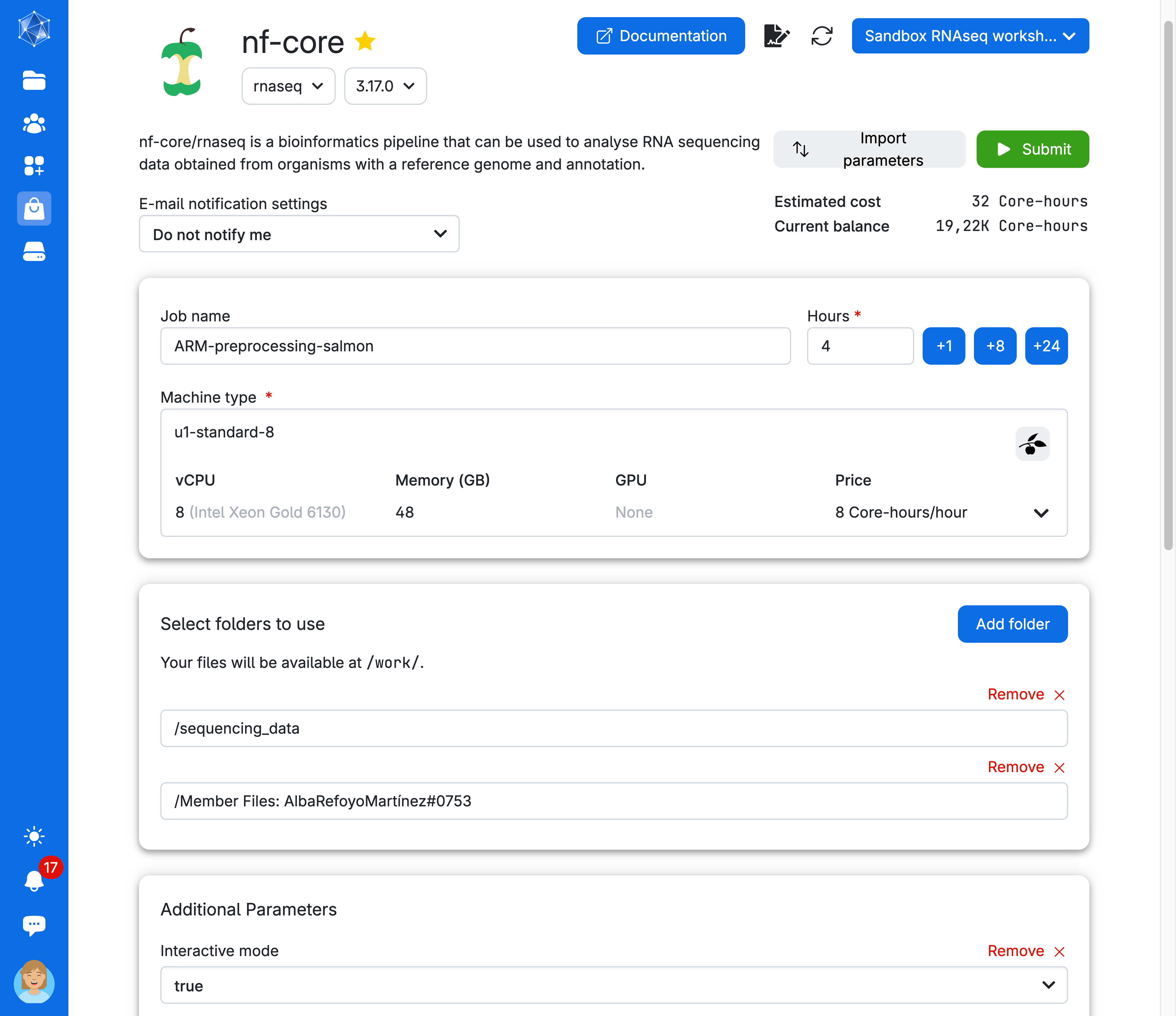
Next, let’s set up the parameters to run the pipeline. Your screen should resemble the image below—please verify it does.
- Assign a job title using your name and the task (e.g., “preprocessing salmon”).
- Select 8 cores and a runtime of 4 hours.
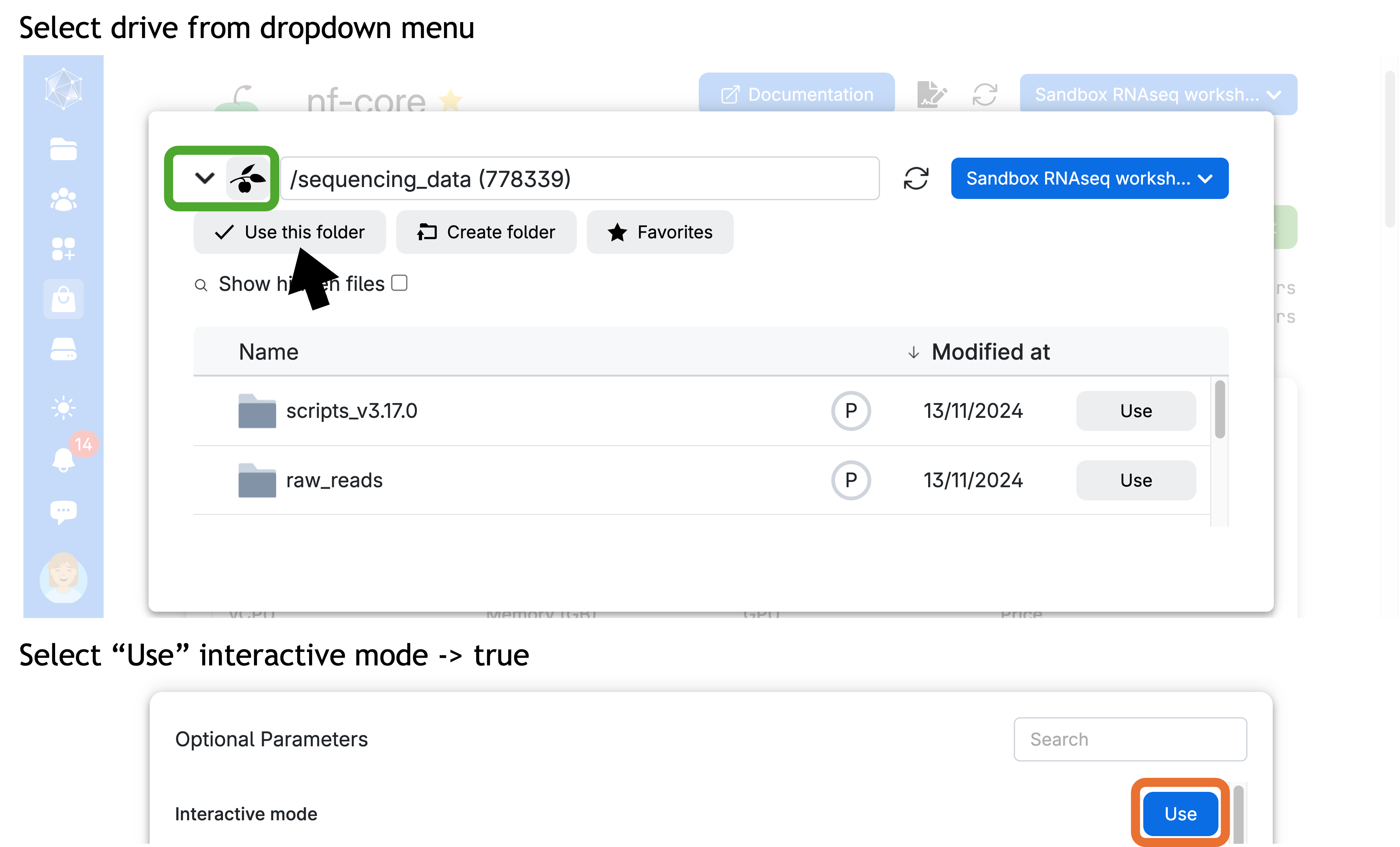
- Choose the sequencing data drive containing the sequencing reads and scripts needed for the pipeline. Please, also add your personal drive—the one with your name (e.g.
Yourname#xxxx). - Enable interactive mode to start a session through the terminal app, allowing us to work with the command line and monitor the progress of the pipeline.
Ensure that the hard-drive icon is labeled “sequencing_data” and you are in the “sandbox_bulkRNAseq” workspace.
You can select the drive by clicking the down arrow (∨) icon to open a dropdown menu.
You are almost ready to run the app! But first, make sure your screen looks like the one shown here. Review the arguments used to run the job: we have specified a Job name, Hours, Machine type, and an additional parameter, Interactive mode.
Now, click the button on the top-right of the screen (submit) to start the job.
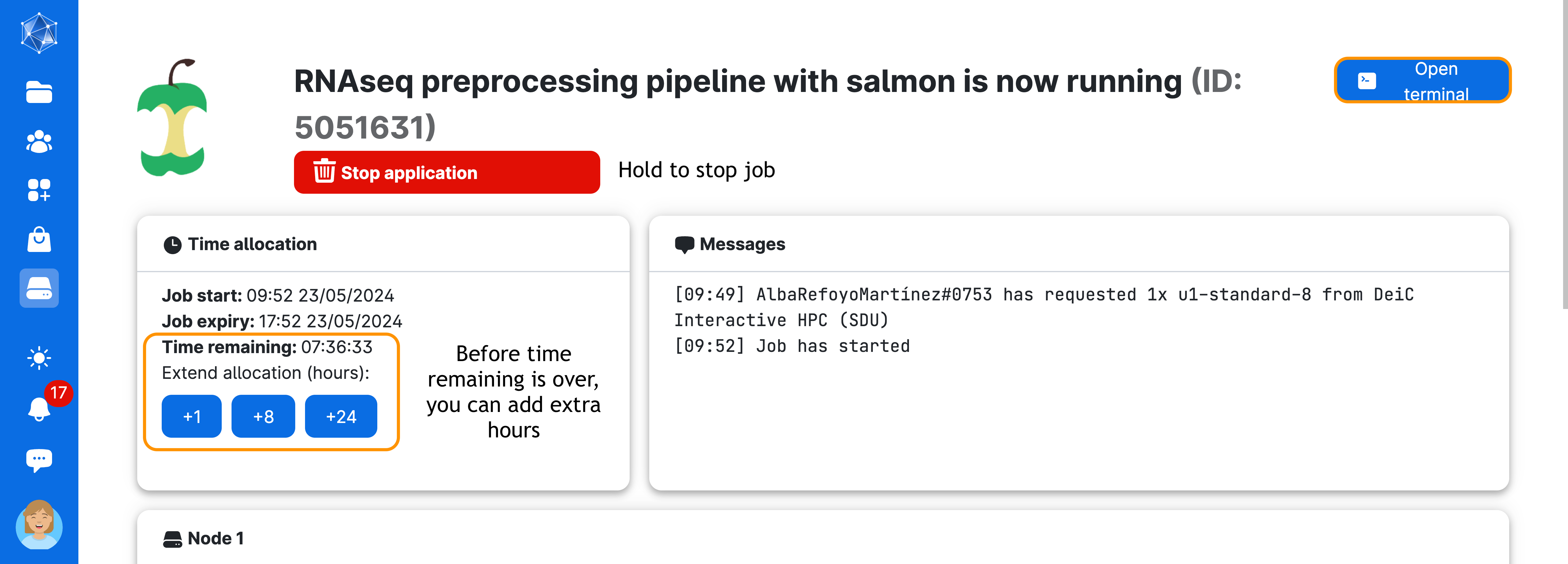
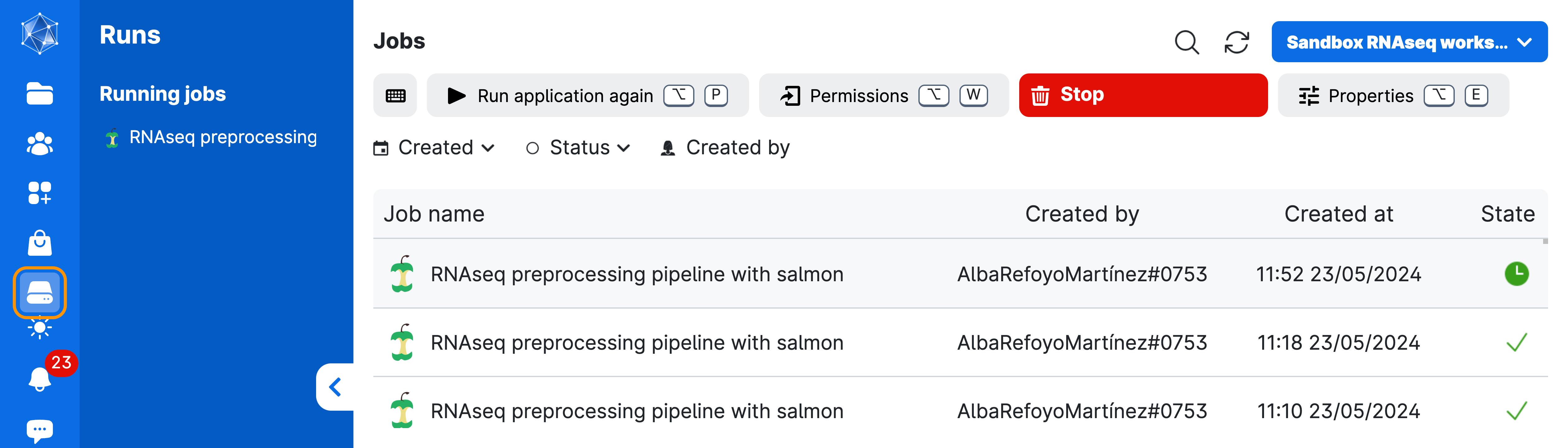
Next, wait until the screen looks like the figure below. This process usually takes a few minutes. You can always come back to this screen via the Runs button in the left menu on UCloud. From there, you can add extra time or stop the app if you no longer need it.
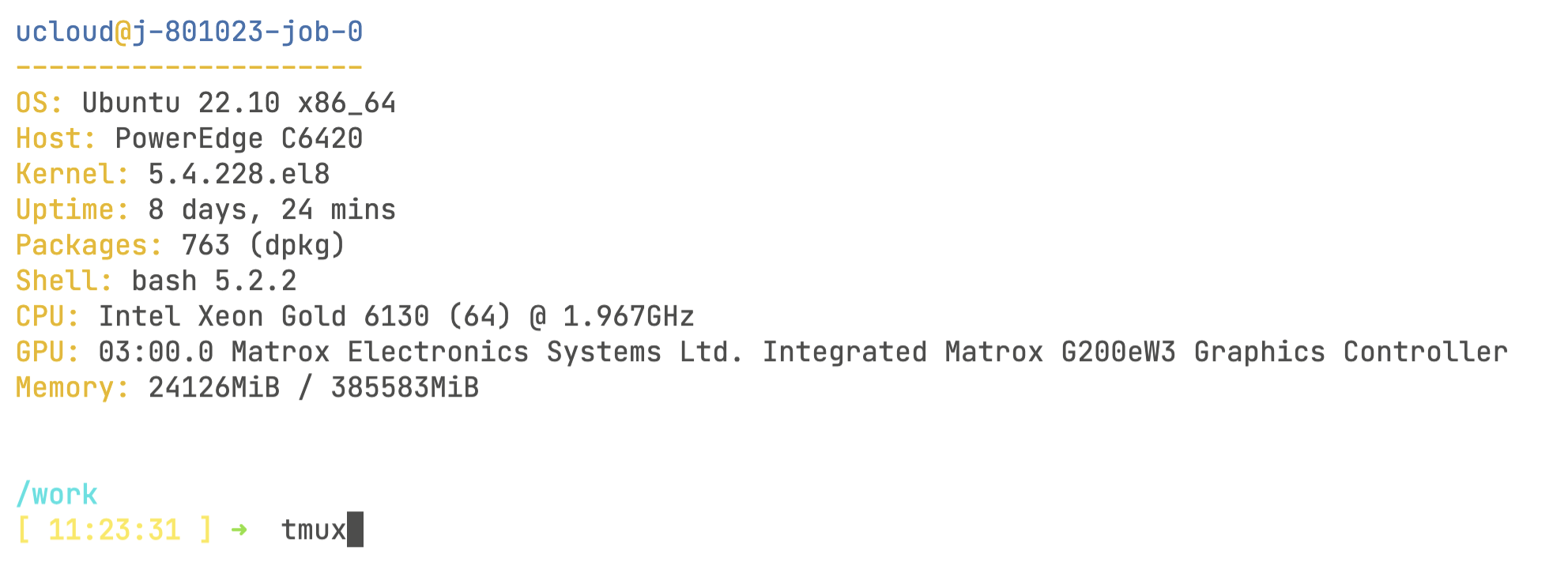
Now, click on Open terminal on the top right-hand side of the screen. You will start the terminal session through your browser! Once inside the terminal, you will need to do one last thing before starting the pipeline:
Terminal
# This will create a session with an automatically assigned numeric name, starting from 0
tmux
# Alternatively,
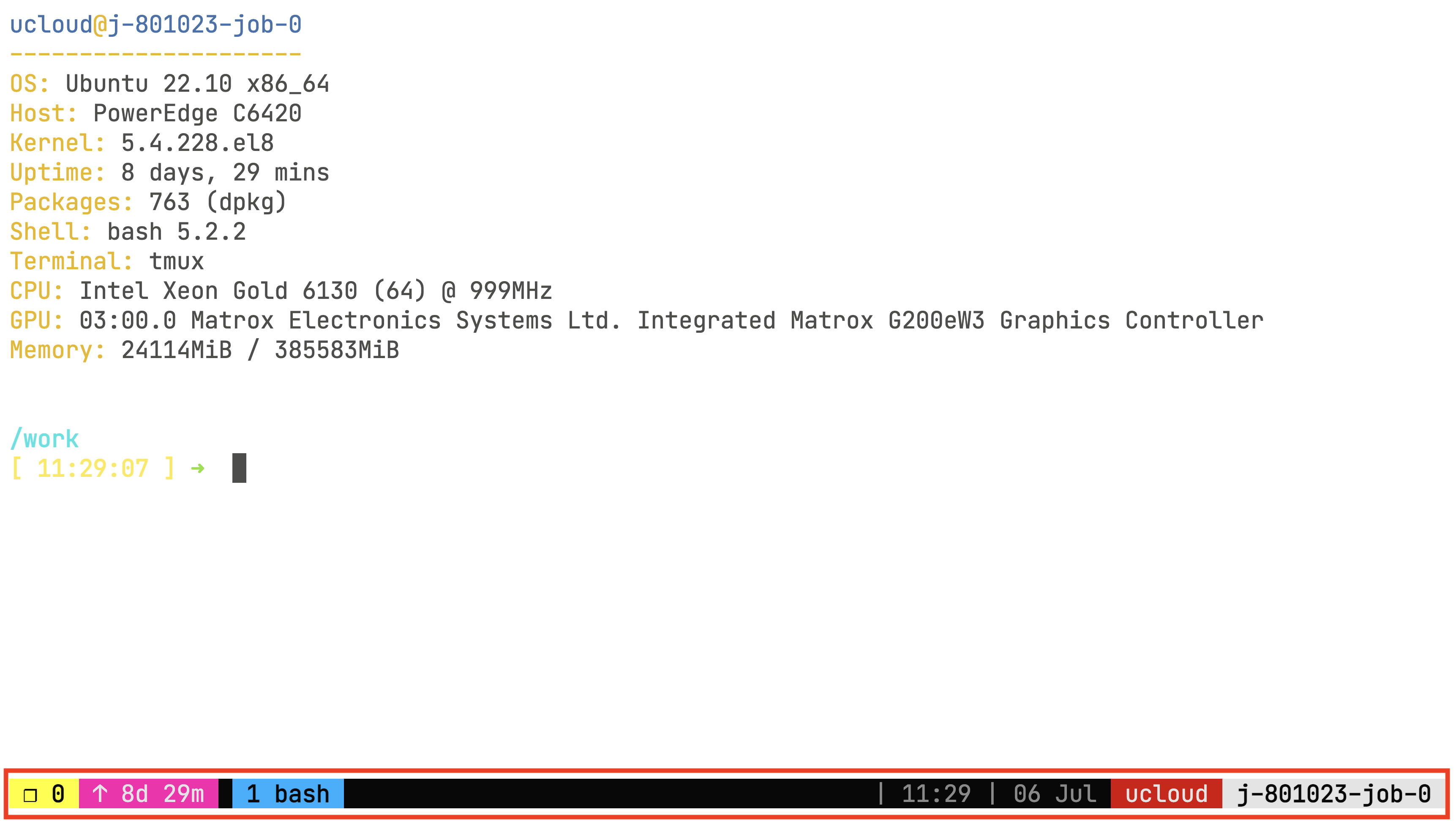
tmux new -s session_nameThe tmux command initiates a persistent virtual command line session. This is crucial because, once the pipeline starts, you won’t be able to monitor its progress if your computer disconnects or enters sleep mode. With tmux, however, you can close your computer, and the session will still be active when you reconnect—unless your requested job time has run out. You’ll know you’re inside the tmux session by checking the bottom of the screen.
If you want to leave the tmux session (and not kill it), you can do so by pressing simultaneously Ctrl and B keys, and then pressing D. Then, you can reconnect to the session using the command:
Terminal
tmux attach -t 0 # abbreviated as tmux a -t0Let’s prepare to run the pipeline. First, navigate to your personal drive—the one with your name (e.g.Yourname#xxxx)—and create a directory to serve as your working directory where all pipeline results will be saved (e.g., nfcore-results). Make sure you’re now located within this new directory. Finally, copy the bash script that runs the Nextflow command into this directory. Check the commands you need to run below:
Terminal
# Change location to your personal drive and create a new directory
# cd /work/<YourName#xxxx>
mkdir nfcore-results
# Make the new dir your current working dir!
cd nfcore-results # You might have named it differently
# Copy our bash script to your directory
cp /work/sequencing_data/scripts_v3.17.0/preprocessing_salmon.sh .
# Make sure you have copied the bash script using the ls command
ls .
# ls is a Linux shell command that lists directory contents of files and directories.
# You will run the pipeline from here as follows BUT not yet! (that's why is commented out)
# bash ./preprocessing_salmon.shOnce copied, review the bash script:
preprocessing_salmon.sh
#!/bin/bash # Specifies the script that should be run in the bash shell
VRNASEQ="3.17.0" # Defines the version of the rnaseq pipeline to use
PARAMSF="/work/sequencing_data/scripts_v3.17.0/nf-params_salmon.json" # Set path to configuration pipeline parameters (e.g. aligner software)
CONFIGF="/work/sequencing_data/scripts_v3.17.0/maxcores.config" # Set path to maximum cores and memory usage (e.g. what you requested on UCloud)
nextflow run nf-core/rnaseq -r $VRNASEQ -params-file $PARAMSF -profile conda -c $CONFIGFThis small bash script initiates the nf-core/rnaseq pipeline. It’s a good practice to save the command used to run the pipeline, and now you’ve copied it into your results folder! You’ll notice extra parameters and configuration settings defined in the script, which we’ll explore shortly. The final line of the script runs the RNA-seq pipeline with the specified version, parameters, Conda profile, and configuration file. We will inspect each part of the Nextflow command below in the section Understanding the pipeline arguments but we will first look at the different input files.
Parameters file
The parameters JSON file is used in nf-core pipelines to define various arguments and software options that can be utilized within the pipeline. You can find a full list of all parameters on the official page: https://nf-co.re/rnaseq/3.17.0/parameters/. Let’s take a look at the different configuration files we’re using.
In the parameters file, we specify that we want to use the pseudoaligner Salmon for this pipeline. The key elements we need to define are the location of the samplesheet, the output directory where Nextflow will save the results, and the paths to the FASTA and GTF files. It is important to use full paths for these locations to ensure that Nextflow can correctly access the required files, regardless of the working directory.
nf-params.json
{
"input": "/work/sequencing_data/raw_reads/samplesheet_v3.17.0.csv",
"outdir": "preprocessing_results_salmon",
"fasta": "/work/sequencing_data/genomic_resources/homo_sapiens/GRCh38/Homo_sapiens.GRCh38.dna_sm.primary_assembly.fa.gz",
"gtf": "/work/sequencing_data/genomic_resources/homo_sapiens/GRCh38/Homo_sapiens.GRCh38.109.MODIFIED.gtf.gz",
"pseudo_aligner": "salmon",
"skip_stringtie": true,
"skip_rseqc": true,
"skip_preseq": true,
"skip_qualimap": true,
"skip_biotype_qc": true,
"skip_bigwig": true,
"skip_deseq2_qc": true,
"skip_bbsplit": true,
"skip_alignment": true,
"extra_salmon_quant_args": "--gcBias"
}--input parameter
The --input parameter points to the samplesheet.csv file that contains all the info regarding our samples.
--outdir parameter
The --outdir parameter indicates where the results of the pipeline will be saved.
--fasta parameter
Path to FASTA reference genome file.
--gtf parameter
Path to GTF annotation file that contains genomic region information.
--pseudo_aligner argument
The --pseudo_aligner argument indicates that we want to use salmon to quantify transcription levels.
Finally, we are skipping several QC and extra steps that we did not explain in the previous lesson. Do not worry if you cannot manage to run the pipeline or you do not have the time, we have prepared a backup folder that contains the results from a traditional alignment + pseudoquantification for you to freely explore! (More about that below).
Samples’ sheet
Let’s quickly explore our samplesheet.csv file (we named it samplesheet_v3.17.0.csv to differentiate it from other formats, as the file may have slightly changed with Nextflow updates—don’t worry about this). Additional details, including descriptions of the columns here
samplesheet_v3.17.0.csv
sample,fastq_1,fastq_2,strandedness
control_3,/work/sequencing_data/raw_reads/Control_3.fastq.gz,,unstranded
control_2,/work/sequencing_data/raw_reads/Control_2.fastq.gz,,unstranded
control_1,/work/sequencing_data/raw_reads/Control_1.fastq.gz,,unstranded
vampirium_3,/work/sequencing_data/raw_reads/Vampirium_3.fastq.gz,,unstranded
vampirium_2,/work/sequencing_data/raw_reads/Vampirium_2.fastq.gz,,unstranded
vampirium_1,/work/sequencing_data/raw_reads/Vampirium_1.fastq.gz,,unstranded
garlicum_3,/work/sequencing_data/raw_reads/Garlicum_3.fastq.gz,,unstranded
garlicum_2,/work/sequencing_data/raw_reads/Garlicum_2.fastq.gz,,unstrandedIt is crucial that the FASTQ paths inside this file match the paths in your job exactly! A good practice is to use the full paths to avoid any discrepancies.
Configuration file
The configuration file ensures that, regardless of how nf-core defines the resources required for each task, the jobs will run with the resources we allocate when submitting the job. You can learn more about configuration parameters here. One of Nextflow’s key strengths is its ability to run on virtually any computational infrastructure, so these settings might differ if you run the workflow on a different machine.
maxcores.config
process {
resourceLimits = [
cpus: 8,
memory: 40.GB,
]
}Understanding the pipeline arguments
Let’s divide the command into different sections.
Nextflow command
nextflow run \
nf-core/rnaseq -r <VERSION>\
-params-file <PARAMS_FILE> \
-profile <conda,docker,arm> \
-c <CONFIG_FILE> \The notation used in the command to run Nextflow is a common practice in bash scripting. It involves defining a variable for the file path (e.g., PARAMSF="/work/sequencing_data/scripts_v3.17.0/nf-params_salmon.json") and then referencing the variable in the command (e.g., -params-file $PARAMSF). This approach enhances the readability and manageability of the script.
You can easily define other paths and reuse the script!
First we have:
nextflow run nf-core/rnaseq -r 3.17.0While usually one would run an nf-core pipeline using nextflow run nf-core/rnaseq and fetch the pipeline remotely, UCloud has installed the pipelines locally. Specifically, we are using the latest version by using the argument -r 3.17.0.
The -params-file argument is another nextflow core argument that allows us to give the nf-core rnaseq pipeline arguments in a json file, instead of creating an excessively long command. Writing the parameters this way allows for better reproducibility since you can reuse the file in the future.
# Set path to configuration pipeline parameters (e.g. aligner software)
PARAMSF="/path/to/params.json"
-params-file $PARAMSFThe next argument defines the -profile. Profiles can give configuration presets for different compute environments. Unfortunately, the UCloud implementation of the nf-core pipelines does not currently allow the use of docker or singularity, which are the recommended profile options. However, UCloud has made sure that there is a working conda environment ready to use!
-profile condaFinally, we include the argument to specify the config file containing the resource settings:
# Set path to maximum cores and memory usage (e.g. what you requested on UCloud)
CONFIGF="/path/to/config.json"
-c $CONFIGFThese are nf-core specific arguments that indicate Nextflow to only use as maximum the number of CPUs and RAM we requested when we submitted the job. We are using 8 cores since it is what we requested on the submission page (e.g. if you submitted a job with 4 CPUs, this will be equal to 4). We are using slightly less RAM than we requested (48Gb) just in case there is a problem with memory overflow.
- Ask your instructors if you have any questions before we start running it.
- You can also check nf-core usage documentation here.
Running the pipeline
We can finally start the run! Type in the command:
Terminal within tmux session
# running command from /work/YourName#xxxx/nfcore-results
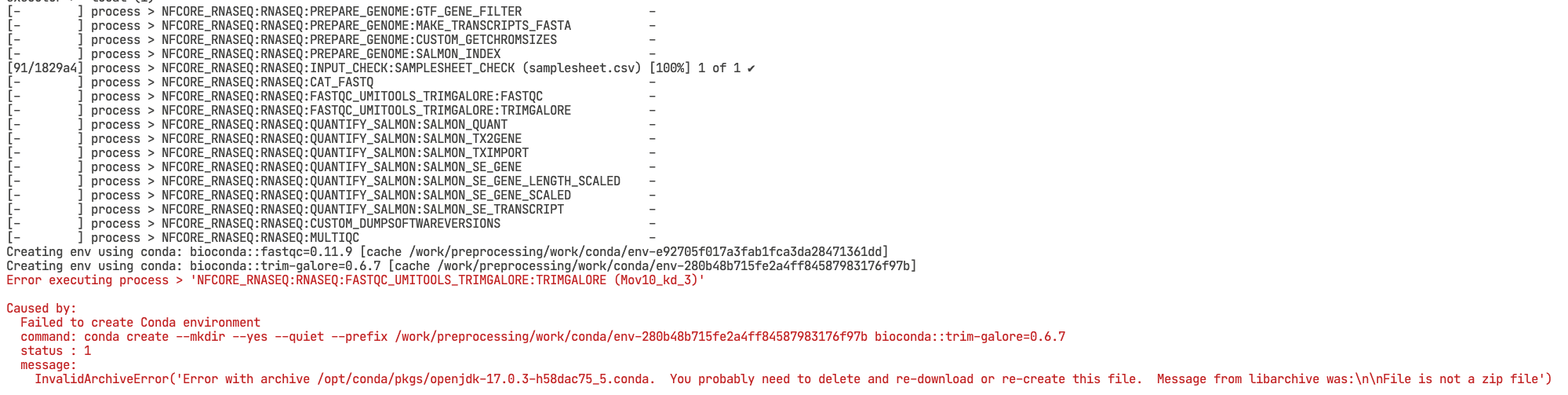
bash ./preprocessing_salmon.sh You should see now a prompt like this, which means that the pipeline started successfully!
Restarting a failed run
When running a nf-core pipeline for the first time, you might encounter some errors, for example, one of your files has an incorrect path, or a program failed to do its job. It is very important to read the error message carefully (try Googling it, as chances are others have encountered the same issue)!
# Error in terminal
Error executing process >
Caused by:
Failed to create create conda environmentOnce you fix the error, it is possible to resume a pipeline instead of restarting the whole workflow. You can do this by adding the -resume argument to the Nextflow command:
nextflow run nf-core/rnaseq -r $VRNASEQ -params-file $PARAMSF -profile conda -c $CONFIGF -resumeStopping the app
Once the pipeline is done (HINT: check your tmux session), go on Runs in UCloud and stop it from using more resources than necessary!
Saved results
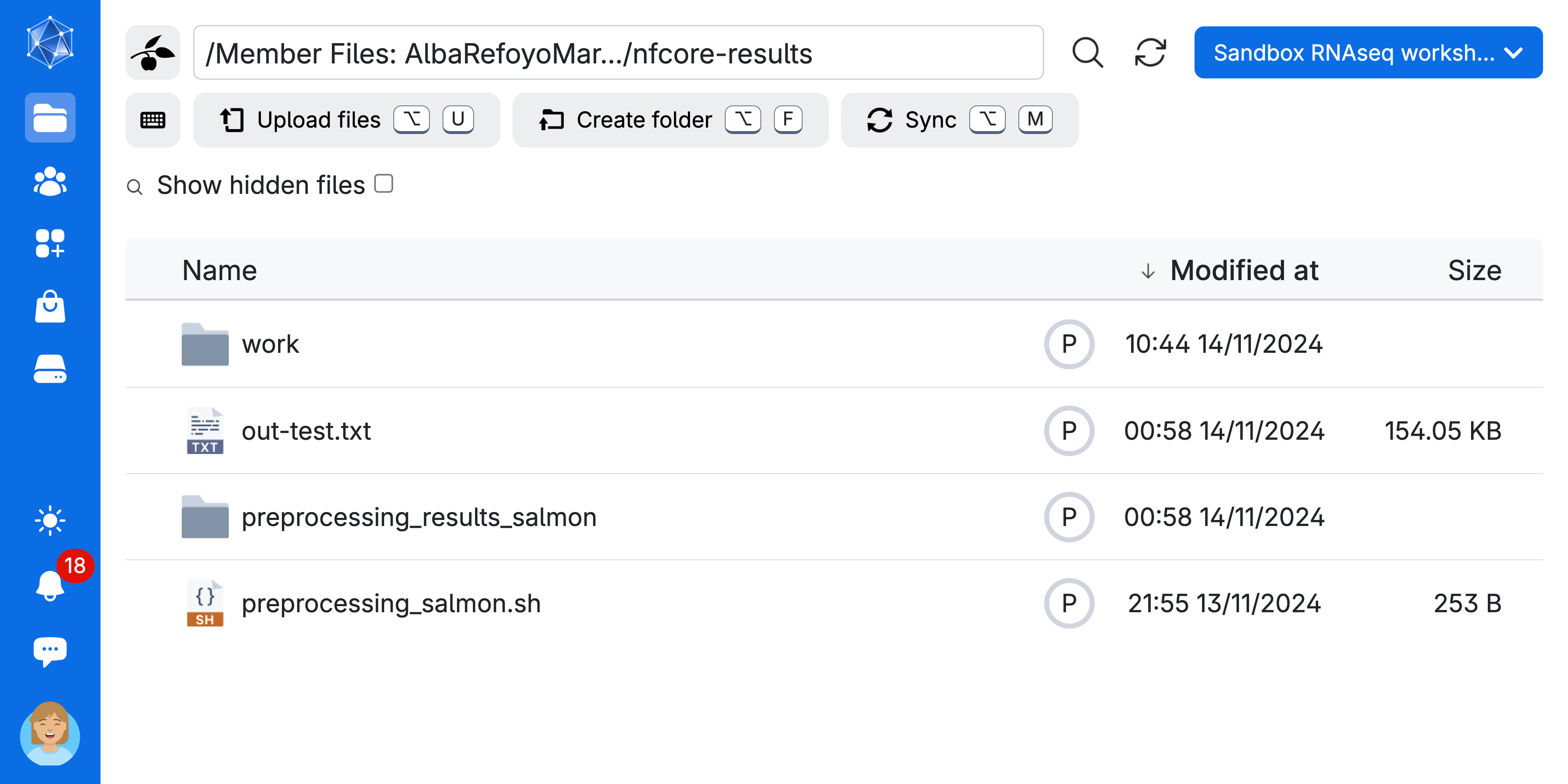
After finishing the job, everything that the pipeline has created will be saved in the directory you created in your personal drive.
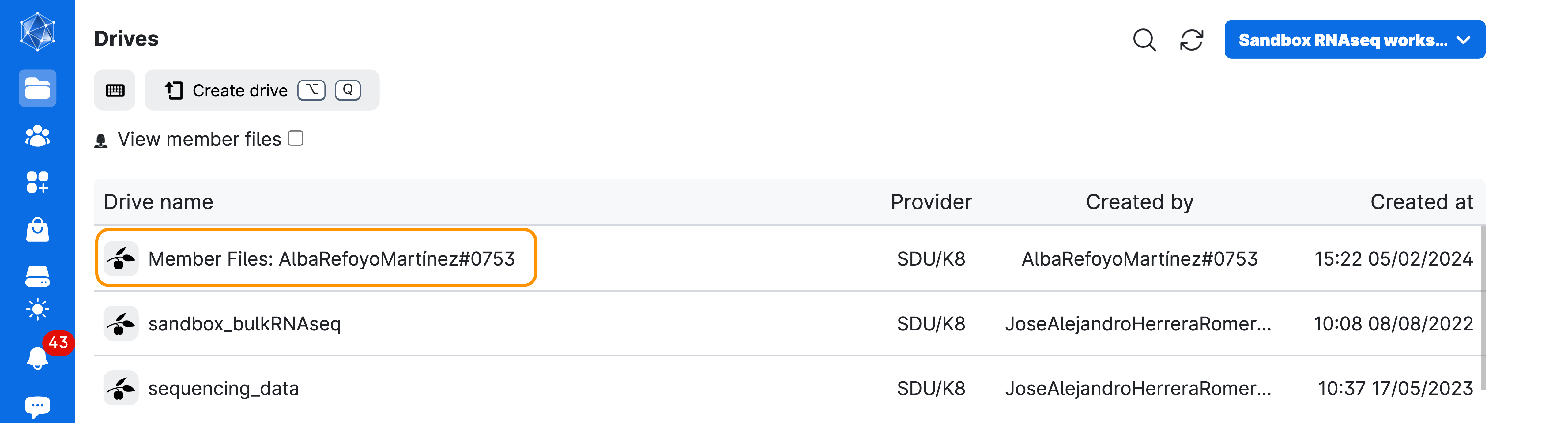
- All generated files will be saved in a volume with your username, which you should be able to find under the menu
Files>Drives>Member Files:YourName#numbers
- You will find the results under the
preprocessing_results_salmonwhich you used as--outdir
A common practice is to remove the work directory once the pipeline completes successfully. This helps save storage space in the long run, as the work directory can accumulate over multiple runs. Feel free to explore the contents to see more details on how the jobs were run, and if there are any failures, you can find additional information there.
You can always find past generated files by looking into the Jobs folder! You will need to remember which app and job name you provided before submitting the job to UCloud. For example,
- Go to
Jobs->nf-core: rnaseq->job_name (RNAseq preprocessing ...)->preprocessing_results_salmon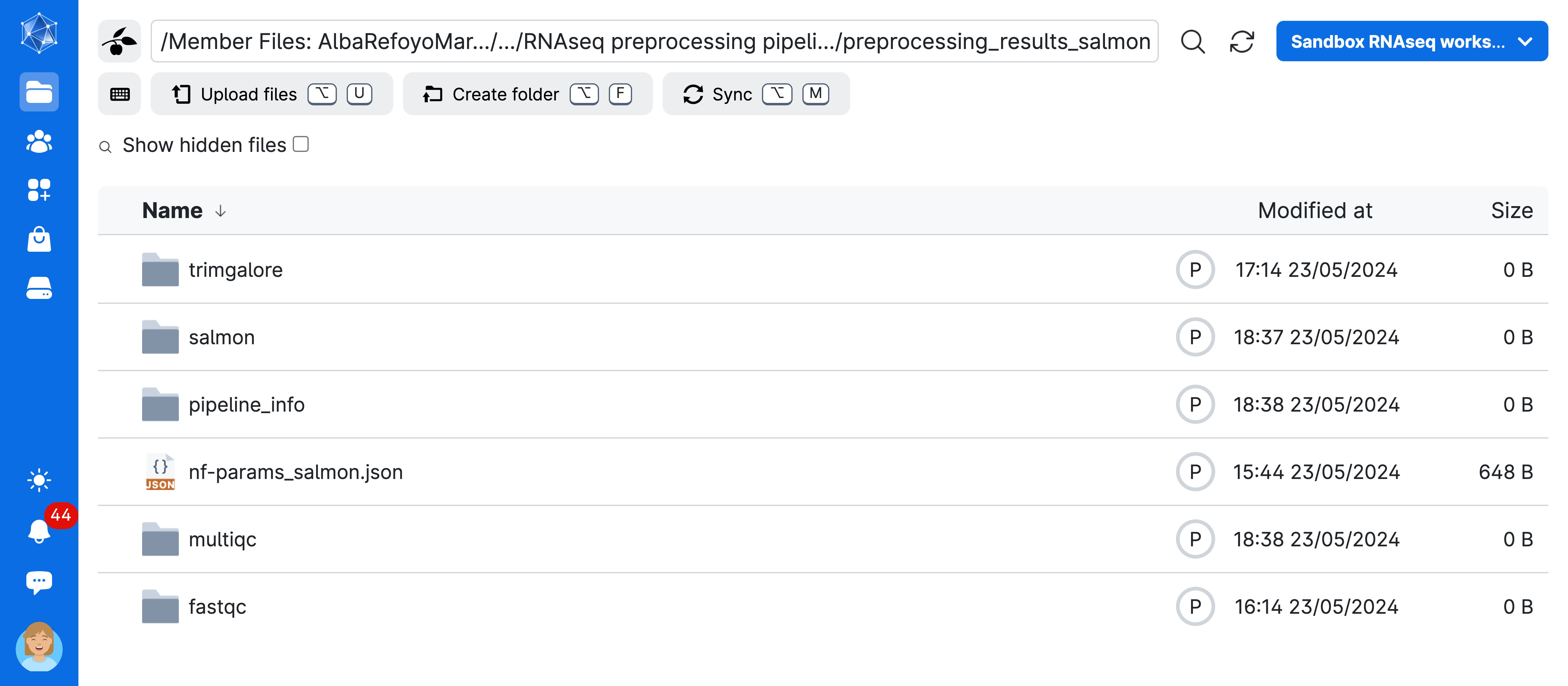
Now you have access to the full results of your pipeline! As explained in the previous lesson, the nf-core rnaseq workflow will create a MultiQC report summarizing several steps into a single html file that is interactive and explorable. In addition, there will be a folder with the results of the individual QC steps as well as the alignment and quantification results. Take your time and check it all out!
Downstream analysis using your results
Optional: If you have successfully run nf-core RNA jobs and want to use the files you generated from your own preprocessing results, don’t forget to choose Use folder when submitting a new job. How?
Go to
Select folders to useandAdd folderby selecting the one containing the pipeline results like shown in the image above. They will be located inMember Files:YourName#numbers->nfcore-results->preprocessing_results_salmon(unless you have moved them elsewhere).