FAIR environments
Computational environments vary significantly from one system to another, encompassing differences in the operating system, installed software, and software package versions. If a research project is moved to a different computer or platform, the analysis might not run or produce consistent results, specially if the software relies on specific configurations or dependencies. Dependencies are libraries or tools that software relies on and can change over time or be poorly documented, leading to hidden variations between setups. Simply knowing a software version might often be insufficient for guaranteeing consistent results across different environments.
If you’re developing your own tools/pipelines, here’s is the hard truth:
- If your code isn’t easy to install, no one will bother using it.
- If you don’t document how to run it, people will be left guessing—and probably give up.
- Lastly, if you don’t carefully control and share your software environment, even you might struggle to get the same results later on.
For research to be reproducible, the original computational environment must be recorded so others can replicate it accurately. This involves making your code easy to install and run by others, document the setup process thoroughly, and carefully manage and share your software environment. There are several methods to achieve this:
- Containerization platforms (e.g., Docker, Singularity): allow the researcher to package their software and dependencies into a standardized container image.
- Virtual Machines (e.g., VirtualBox): can share an entire virtualized computing environment (OS, software and dependencies).
VMs simulate a complete operating system, including virtualized CPU, RAM, and storage. They are highly isolated, providing a separate environment for each VM. While VMs offer excellent isolation, they can be slow due to the overhead of simulating hardware and have large images that are cumbersome to share. VMs also require data transfer over the network to interact with the host system, which can be inefficient.
- Environment package managers: provide an isolated environment with specific packages and dependencies that can be installed without affecting the system-wide configuration. These environments are particularly useful for managing conflicting dependencies and ensuring reproducibility. Configuration files can automate the setup of the computational environment:
- conda or mamba: allows users to export environment specifications (software and dependencies) to YAML files enabling easy recreation of the environment on another system. Conda disadvantage: long-term reproducibility.
- Python
virtualenvorpipenv: is a tool for creating isolated environments to manage dependencies specific to a project. - requirements.txt: may contain commands for installing packages (such as pip for Python packages or apt-get for system-level dependencies), configuring system settings, and setting environment variables. Package managers can be used to install, upgrade and manage packages.
- R’s
renv: The ‘renv’ package creates isolated environments in R.
- Environment descriptors: refer to various types of metadata or attributes that provide information about the computational environment in which software or applications are running (e.g.
__version__,sessionInfo()). Check more useful commands here.
- Kernel mechanisms such as chroot, namespaces and control groups can also be used for process isolation and resource management.
- Chroot (filesystem isolation): changes the directory for a process, isolating its filesystem view from the rest of the system so that the process can only access files within a specified directory tree.
- Namespace (process isolation): isolates processes from each other by creating separate namespaces for various resources such as process IDs, network interfaces, and filesystems. Processes within the same namespace can interact with each other, but they are isolated from processes in other namespaces.
- Control groups (resource management): manage and limit the amount of system resources that processes can use which ensures that they cannot exceed their allocation resources and helps maintaining the overall system performance and stability.
While package managers are very easy to use and share across different systems, and are lightweight and efficient, offering fast start-up times, containers provide a full environment isolation (including the operating system) which ensures consistent behavior across different systems. Check the table below for a full overview.
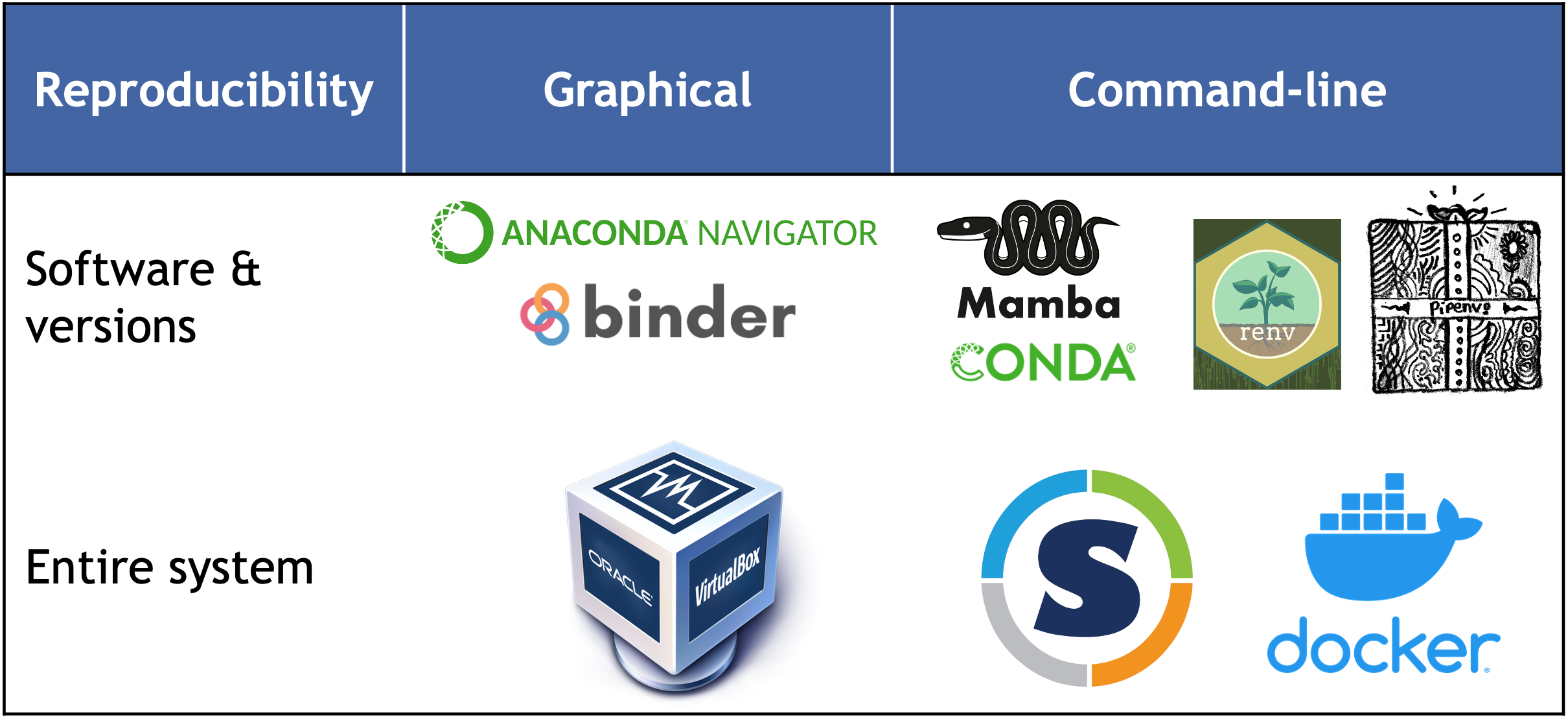
Recording and sharing the computational environment is essential for ensuring reproducibility and transparency. Below, we will explore two tools that can help with this: mamba, a package manager, and Docker, a container system. We will explain the differences between them and provide guidance on choosing the right tool for your specific scenario.
Package managers
Mamba is a reimplementation of the Conda package manager in C++. While our focus will be on Mamba, it’s important to note that it maintains compatibility with Conda by using the same command-line parser, package installation and uninstallation code, and transaction verification routines.
Mamba uses software installation specifications that are maintained by extensive communities of developers, organized into channels, which serve as software repositories. For example, the “bioconda” channel specializes in bioinformatics tools, while “conda-forge” covers a broad range of data science packages.
As previously mentioned, mamba is a newer and faster implementation. The two commands can be used interchangeable (for most tasks). If you use Conda, you should still complete the exercises, as you’ll gain experience with both tools. For more information on their ecosystem and advantages here.
Mamba allows you to create different software environments, where multiple package version can co-exit on your system.
Follow mamba instructions to install it. Let’s also include bioconda and conda-forge channels which will come very handy.
conda config --add channels defaults; conda config --add channels bioconda; conda config --add channels conda-forgeNow you are set to create your first environment. Follow these steps:
- Create a new environment named myenv
- Install the following packages in
myenv:bowtie2,numpy=1.26.4,matplotlib=3.8.3 - Check the environments available
- Load/activate the environment
- Check which
pythonexecutable is being used and that bowtie2 is installed. - Deactivate the environment
Here are some of the commands you need for the exercise.
mamba create -n <ENV-NAME>
mamba install --channel <CHANNEL-NAME> --name <ENV-NAME>
mamba env list
# mamba init
mamba activate <ENV-NAME>
mamba deactivate - The syntax to create a new environment is:
mamba create --name myenv - Example “bowtie2”: Go to anaconda.org and search for “bowtie2” to confirm it is available through Mamba and which software channel it is provided from. You will find that it is available via the “bioconda” channel: https://anaconda.org/bioconda/bowtie2. The syntax to install packages is:
mamba install --channel <CHANNEL-NAME> --name <ENV-NAME> <SOFTWARE-NAME>
mamba install --name myenv --channel bioconda bowtie2=2.5.3 "matplotlib=3.8.3" "numpy=1.26.4"
Do the same with the others. 3. To see al environments available mamba env list. There will be a “*” showi8ng the one is activated. 4. Load the environment mamba activate myenv. 5. which python -> should print the one in the environment that is active (path similar to /home/mambaforge/envs/myenv/bin/python). bowtie2 --help 6. Conda deactivate
If you have different environments set up for various projects, you can switch between them or run commands directly within a specific environment using:
mamba run -n <ENV-NAME> python myscript.pyIf you need to activate an environment in a shell script that will be submitted to SLURM, you must first source Mamba’s configuration file. For instance, to load the myenv environment we created, the script would include the following code:
# Always add these two commands to your scripts
eval "$(conda shell.bash hook)"
source $CONDA_PREFIX/etc/profile.d/mamba.sh
# then you can activate the environment
mamba activate myenvWhen jobs are submitted to SLURM, they run in a non-interactive shell where Mamba isn’t automatically set up. By running the source command, you ensure that Mamba’s activate function is available. It’s important to remember that even if the environment is loaded on the login node, the scripts will execute on a different machine (one of the compute nodes). Therefore, always include the command to load the Mamba environment in your SLURM submission scripts.
Containers
Essentially, a container is a self-contained, lightweight package that includes everything needed to run a specific application—such as the operating system, libraries, and the application code itself. Containers operate independently of the host system, which allows them to run the same software across various environments without any conflicts or interference. This isolation ensures that researchers can consistently execute their code on different systems and platforms, without worrying about dependency issues or conflicts with other software on the host machine.
The most significant difference is at the permission level required to run them. Docker containers operate as root by default, giving them full access to the host system. While this can be useful in certain situations, it also poses security risks, especially in multi-user environments. In contrast, Singularity containers run as non-root users by default, enhancing security and preventing unauthorized access to the host system.
- Docker is ideal for building and distributing software across different operating systems
- Singularity is designed for HPC environments and offers high performance without needing root access
In the following sections, we’ll cover how to retrieve environment information, utilize containers, and automate environment setup to improve reproducibility.
Singularity on a remote server
While you can build your own Singularity images, many popular software packages already have pre-built images available from public repositories. The two repositories you’ll most likely use or hear about are:
# You will only need to vagrant init once
export VM=sylabs/singularity-3.0-ubuntu-bionic64 && \
vagrant init $VM && \
vagrant up && \
vagrant ssh- We recommend using the pre-installed version provided by your system administrators if you’re working on a shared system. If you’re working on your own computer, you can install the necessary software using Mamba.
- They might host different versions of the same software, so it’s worth checking both to find the version you need.
- To download a software container from public repositories, use the
singularity pullcommand. - To execute a command within the software container, use the
singularity runcommand. - Good practice: create a directory to save all singularity images together.
.sifis the standard extension for the images.
Download a singularity image from one of the two repositories listed above (choose a software like bcftools, bedtools, bowtie2, seqkit…) and run the --help command. This command displays the help documentation of the program, verifying that our image is functioning correctly and includes the intended software.
# create a directory for our singularity images
mkdir images
# download the image
singularity pull images/bowtie2-2.5.4.sif https://depot.galaxyproject.org/singularity/bowtie2%3A2.5.4--he20e202_2
# run the image: singularity run <PATH-TO-IMAGE> <YOUR COMMANDS>
singularity run images/bowtie2-2.5.4.sif bowtie2 --helpSources
- Anaconda for searching Mamba/conda packages
- Bioconda for installing software package related to biomedical research
- Conda cheat sheet
- faircookbook worflows
- Docker
- Docker get-started
- The turing way - reproducible research
Find pre-built singularity images:
Other training resources: - The turing way - reproducible research - HPC intro by Cambridge - Highly recommend Reproducible Research II: Practices and Tools for Managing Computations and Data by members of France Université Numérique.