7. Storing and sharing biodata
⏰ Time Estimation: X minutes
💬 Learning Objectives:
- Repositories for managing biological data
- Archive your GitHub data analysis repositories
While platforms like GitHub excel in version control and collaborative coding, repositories specialize in archiving and sharing scientific data (e.g. Zenodo), ensure long-term accessibility for the global research community.
What to archive and how?
A framework for reproducibility in computational research can generally be divided into three key, though sometimes overlapping, categories:
- Readable Components: This includes elements such as literature reviews, code documentation, data documentation, and workflow descriptions outlining how the code interacts with the data.
- Executable Components: These are the actual code, scripts, and software that need to be compiled and run to reproduce results.
- Interpretable Components: This refers to the data itself—raw or processed—that the code and scripts work on.
Researchers are typically more accustomed to archiving readable components, such as papers or data documentation, compared to executable components like scripts and code. However, for research to be fully reproducible, it is crucial that all key components, including executable ones, are properly archived.
When choosing an archival solution, it’s important to recognize that there is no one-size-fits-all option. Several factors must be considered, including data size, format requirements, licensing conditions, cost, and tools for data attribution and citation. Each of these features plays a crucial role in selecting the most suitable archive for your needs.
Data Repositories and Archives
Specialized repositories and archives securely store, curate, and disseminate scientific data, ensuring long-term preservation, transparency, and citability of research findings through standardized formats and rigorous curation processes.
Importance of archiving scientific data
- Long-term accessibility and preservation: Ensures data remains accessible for future researchers.
- Enhanced visibility and attribution: Unique identifiers like DOIs enable citation of datasets, enhancing visibility and proper attribution.
- Improved dataset discoverability and interpretability: Comprehensive metadata, including methodology and experimental details, facilitates understanding and usability by other researchers.
- Promotion of Transparency, Reproducibility, and Research Integrity: Mandatory data deposition fosters transparency and upholds research integrity.
- Amplification of Research Impact and Contribution: Archiving data elevates research quality and extends its impact within the scientific community.
- Fulfilling Scholarly Obligations: Compliance with requirements set by scientific journals and funding agencies ensures adherence to scholarly standards.
Check the registry of research data repositories,re3data.org for a full overview. You can browse by subject if you are looking within a specific field.
There are two types of repositories:
- General repositories: relevant to a wide range of disciplines (e.g. Zenodo).
- Domain-specific: repositories are customized for specific fields, providing specialized curation and context-specific features (e.g. ENA, GEO, Annotare, etc.)
- European Nucleotide Archive (ENA)
- NCBI Gene Expression Omnibus (GEO)
- Sequence Read Archive (SRA)
- Protein Data Bank (PDB)
- Proteomics Identifications Database (PRIDE)
- Universal Protein Resource (UniProt) SPIN
- ArrayExpress
- EBI Metagenomics (MGnify)
- PhysioNet
- Functional Annotation of Animal Genomes (FAANG) Data Repository
For more data repositories, please refer to the links provided below to find the appropriate repository for your data:
Your institution might as well have its repositories such as ERDA (Electronic research data archive at the University of Copenhagen).
Domain-specific repositories
This tailored approach ensures alignment with standards and maximizes the utility and impact of research findings. By catering to particular research areas, these repositories offer researchers a more focused audience, deeper domain expertise, and increased visibility within their specific research community.
Explore some examples of NGS data repositories below:
ENA: hosted by the European Bioinformatics Institute (EBI), provides researchers with a platform to deposit and access nucleotide sequences along with associated metadata, ensuring data preservation and contextualization. ENA adheres to community standards and guidelines for data submission, including those established by the International Nucleotide Sequence Database Collaboration (INSDC).
GEO (Gene Expression Omnibus): curated by the National Center for Biotechnology Information (NCBI), serves as a specialized repository for high-throughput functional genomic data sets, particularly gene expression data across diverse biological conditions and experimental designs. Researchers can easily deposit and access a variety of genomic data, fostering data transparency and reproducibility within the scientific community. GEO assigns unique accession numbers to each dataset, ensuring traceability and proper citation in research publications.
ArrayExpress/Annotare: hosted by the European Bioinformatics Institute (EBI), is a specialized repository tailored for storing and submitting functional genomics experiments for high-throughput sequencing data. It offers researchers a platform to upload experimental data along with comprehensive metadata ensuring preservation and contextualization. Annotare provides a curated environment aligned with the standards and practices of the field. This specialization enhances data discoverability, promotes collaboration, and facilitates deeper insights into the functional aspects of the genome.
The repositories mentioned earlier adhere to established community standards for data submission and sharing in genomics research such as:
- MIAME (Minimum Information About a Microarray Experiment): These guidelines ensure comprehensive and standardized reporting of microarray experiments.
- MIxS (Minimum Information about a high-throughput SeQuencing Experiment): MIxS standards, developed by the Genomic Standards Consortium, ensure consistent reporting of metadata for high-throughput sequencing experiments.
- Sequence Read Archive (SRA) Submission Guidelines: They include requirements for data formatting, metadata inclusion, and quality control.
- Community-Specific Standards designed to ensure that submitted data meets the specific requirements and expectations of the field.
By adhering to standards, repositories ensure that submitted data is high quality, well-documented, and compliant with community best practices, promoting data discovery, reproducibility, and interoperability within the scientific community.
Following all the recommendations in this course makes it straightforward to provide the necessary documentation and information for these repositories. For instance, repositories specific to NGS data will require the raw FASTQ files, sample metadata, and protocols as well as final pre-processing results (for instance, read count matrices in BED files).
Software and computations archives
Keep in mind that data repositories are not intended for downstream analysis data and associated code. However, you should already have those versions controlled by GitHub, which eliminates any concerns. You can then archive such repositories in a general repository like Zenodo.
Archives for software source code are essential for long-term accessibility and reproducibility and are becoming very popular. Check Software Heritage if you are developing software.
General repositories
There are plenty of general archiving repositories. We recommend to check the Longwood Research Data management website at Harvard for a quick overview. Some of the most well-known are:
- Dataverse
- Dryad
- figshare
- Open Science Framework (OSF)
- Zenodo
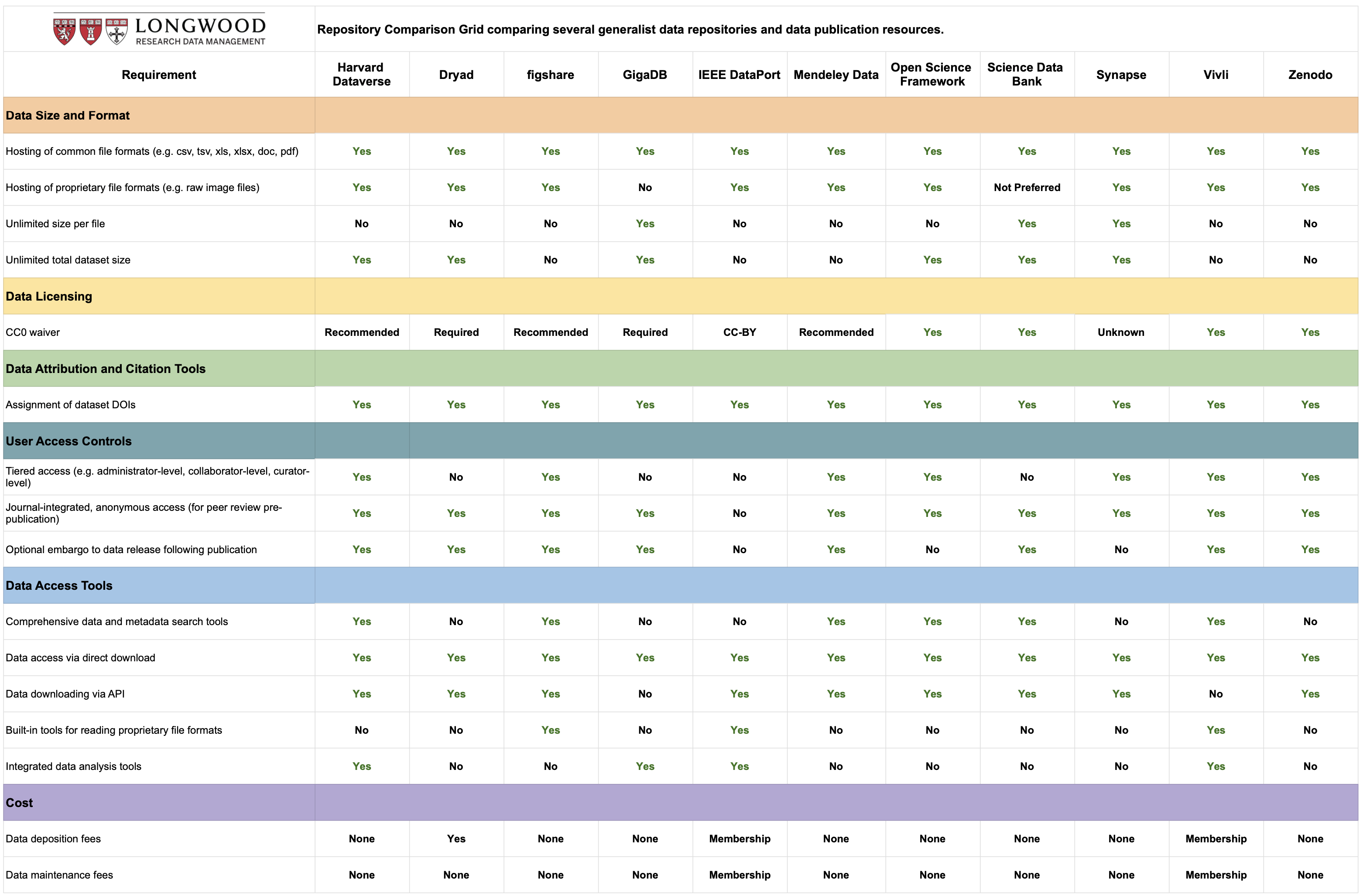
We will be using Zenodo for our practical workshop. However, please review the table provided by the Longwood, Harvard Biomedical Data Management team, which outlines the differences between various repositories.
Zenodo
Zenodo is one of the widely used repositories for a variety of research outputs. It is an open-access digital platform supported by the European Organization for Nuclear Research (CERN) and the European Commission. It caters to various research outputs, including datasets, papers, software, and multimedia files, making it a valuable resource for researchers worldwide. With its user-friendly platform, researchers can easily upload, share, and preserve their research data. Each deposited item receives a unique Digital Object Identifier (DOI), ensuring citability and long-term accessibility. Additionally, Zenodo offers robust metadata capabilities for enriching submissions with contextual information. Moreover, researchers can link their GitHub accounts to Zenodo, simplifying the process of archiving the GitHub repository releases for long-term accessibility and citation.
Once your accounts are linked, creating a Zenodo archive becomes as straightforward as tagging a release in your GitHub repository. Zenodo automatically detects the release and generates a corresponding archive, complete with a unique Digital Object Identifier (DOI) for citable reference. Therefore, before submitting your work to a journal, link your data analysis repository to Zenodo, obtain a DOI, and cite it in your manuscript which enhances reproducibility in research.
Step-by-Step Setup Guide
Check the practical material where we demonstrate how to link Zenodo and Github (see Exercise 6 in the practical material).
Wrap up
In this concluding lesson, we’ve covered the process of submitting your data to a domain-specific repository and archiving your data analysis GitHub repositories in Zenodo. By applying the lessons from this workshop, you’ll significantly enhance the FAIRness of your data and improve its organization for future use. These benefits extend beyond yourself to your teammates, group leader, and the wider scientific community.
We hope that you found this workshop useful. If you would like to leave us some comments or suggestions, feel free to contact us.