Welcome to GWAS with the Genomics Sandbox
You are expected to make sure you can sign in to UCloud, SDU’s HPC platform on which we will be running this course. All data, assignments, and tools will be provided on UCloud. Please use your university ID to sign in (instructions below). If you run into problems, please write us (respond to the email that got you to this page).
Access Sandbox resources
Our first choice is to provide all the training materials, tutorials, and tools as interactive apps on UCloud, the supercomputer located at the University of Southern Denmark. Anyone using these resources needs the following:
- a Danish university ID so you can sign on to UCloud via WAYF1.
basic ability to navigate in Linux/R/Jupyter. You don’t need to be an expert, but it is beyond our ambitions (and course material) to teach you how to code from zero and how to run analyses simultaneously. We recommend a basic R or Python course before diving in.
For workshop participants: Use our invite link to the correct UCloud workspace that will be shared on the day of the workshop. This way, we can provide you with compute resources for the active sessions of the workshop2. Click the link below to accept the invitation (after accessing UCloud for the first time).
Invite link to UCloud workspace
- Download the slides
- EXTRA EXERCISE - GWAS4
You can download an extra notebook and data with population structure and effect of LD on that. Upload the files through jupyterlab, and copy the data in the data folder and the notebook in the notebooks folder:
- EXTRA EXERCISE - LDAK
A notebook with an analysis using LDAK for preprocessing and some of the implemented association testings and heritability estimates
- Course evaluation survey - The Novo Nordisk Foundation funds the Sandbox project and is interested in the outcomes of our training activities, so we really appreciate your responses!
Discussion and feedback
We hope you enjoyed the workshop. If you have broader questions, suggestions, or concerns, now is the time to raise them! Remember that you can check out longer versions of our tutorials as well as other topics and tools in each of the Sandbox modules. All of our open-source materials are available on our Github page and can be used on any computing cluster! We regularly run workshops on a variety of health data science topics that you can also check out (follow our news here).
Download or copy your data
You can move all your data into your own personal workspace on UCloud, and also download all data and nortebooks on your computer.
Copy data on UCloud
If you transfer the data to you UCloud’s workspace, you can at any time run the course material with the Genomics Sandbox application and use the folders with your own Notebooks and datasets.
Go on the left-side toolbar and click on the file browser.

Be sure you are in the Sandbox_workshop workspace (green circle). You should be able to see your personal folder whose name starts with Member Files (red circle).
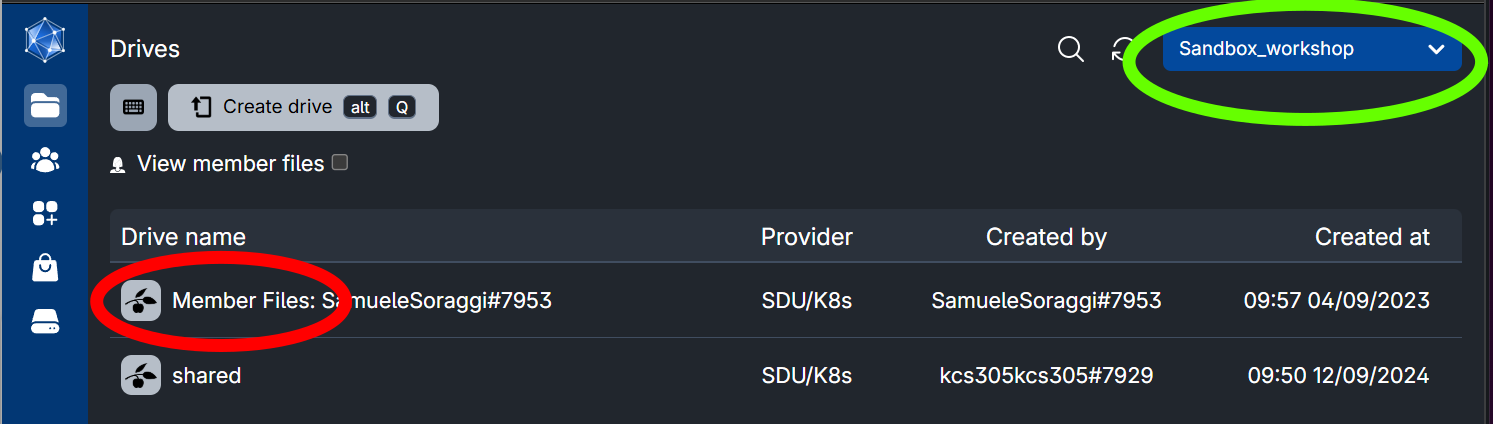
Get into your personal folder and find the folder containing all your course material. Right-click on it, and choose Copy to…
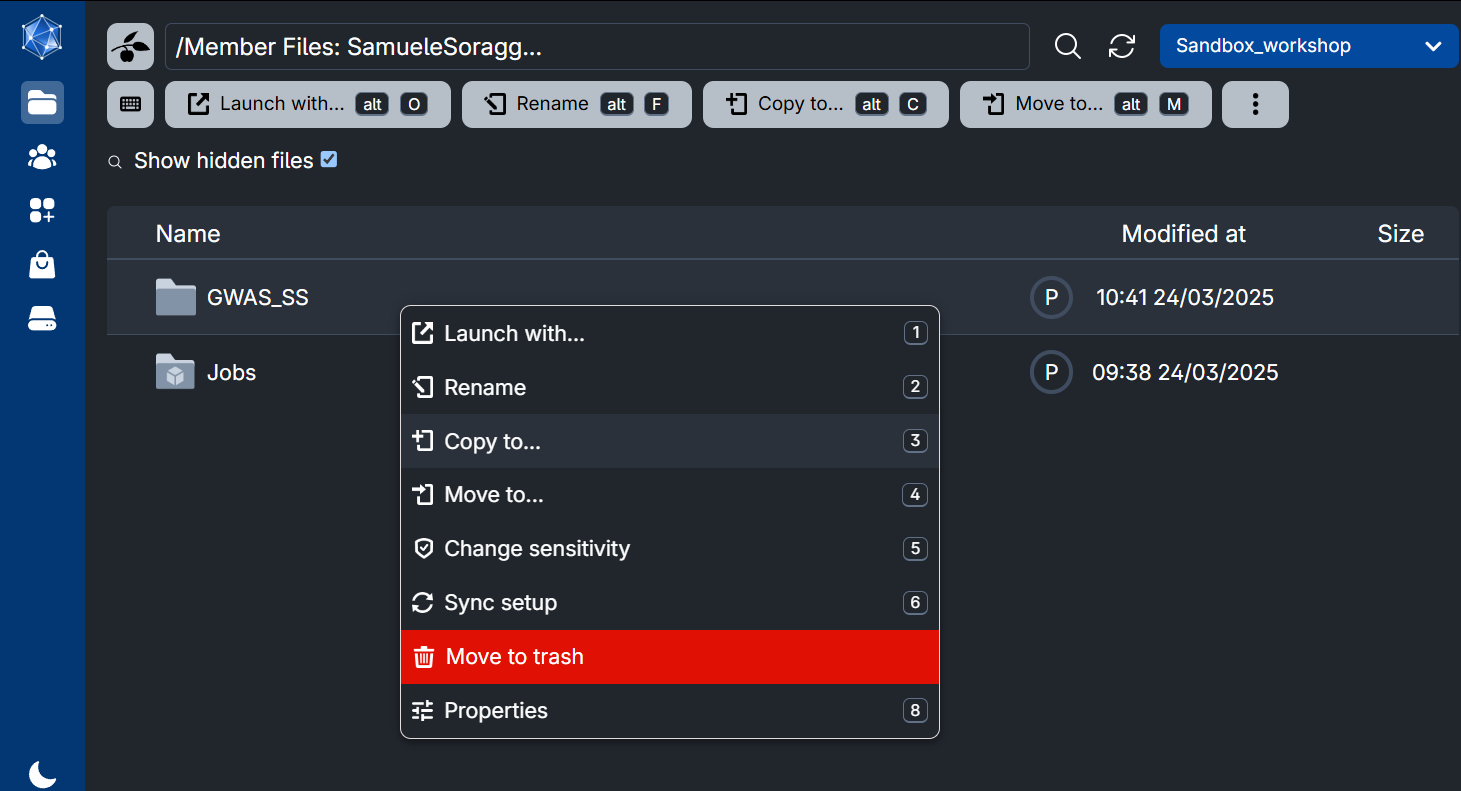
In the browsing window which opens, choose your own workspace My workspace (green circle). You should be already inside your Home folder, otherwise you can find it with the small arrow (red circle). Now click on Use this folder (blue circle). The data is copied.
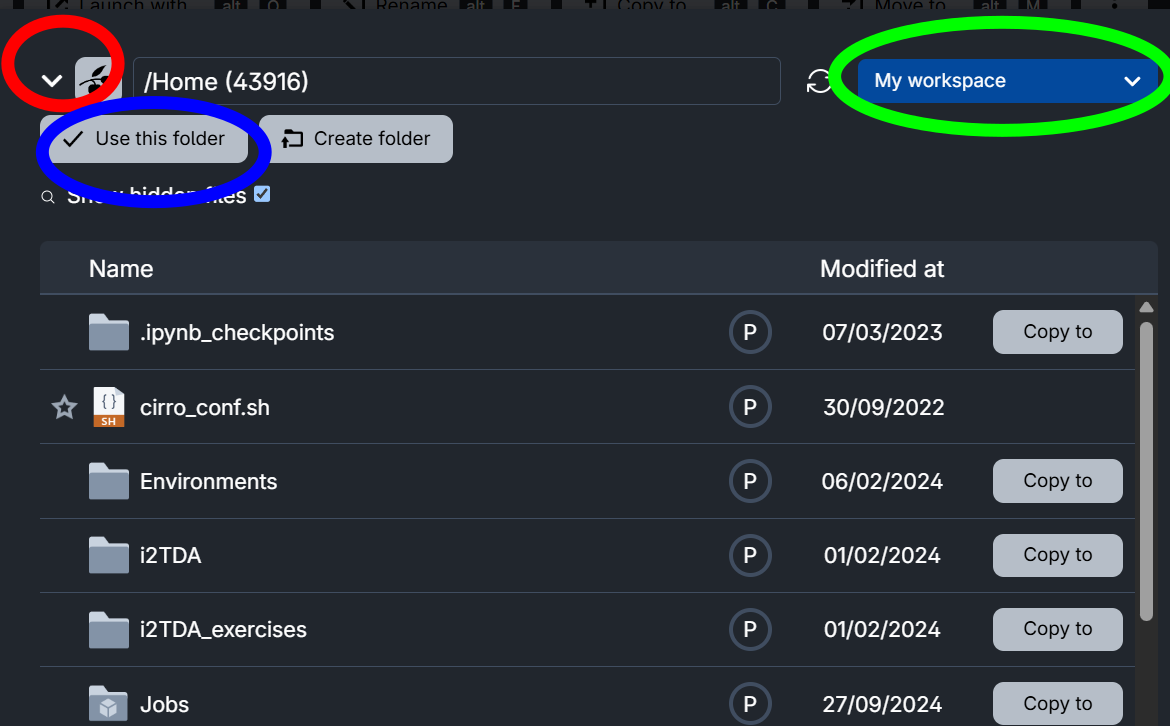
Next time, when you launch the genomics sandbox, use
Add Folderand select the Notebooks and Data folder from your personal workspace.
Download data from UCloud
To download the data and notebooks, you first need to compress it in a zip file.
Launch the genomics sandbox with the data and notebooks as added folders.
When you open the graphical interface, open a new terminal (green circle)

Run the following command to go into the
/workfolder, where the material is loaded.cd /workCreate a compressed version of your folders. It takes some time, so wait for it.
zip -r GWAS.zip Data NotebooksRight-click on the file
GWAS.zip, and choose download. It will be transferred to your download folder!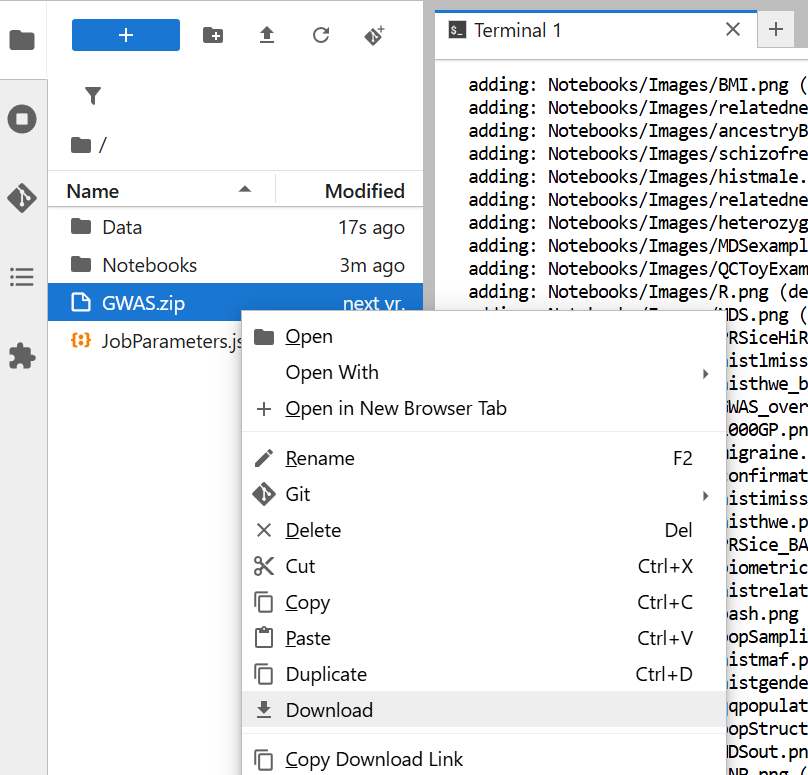
Footnotes
Other institutions (e.g. hospitals, libraries, …) can log on through WAYF. See all institutions here.↩︎
To use Sandbox materials outside of the workshop: remember that each new user has hundreds of hours of free computing credit and around 50GB of free storage, which can be used to run any UCloud software. If you run out of credit (which takes a long time) you’ll need to check with the local DeiC office at your university about how to request compute hours on UCloud. Contact us at the Sandbox if you need help or want more information.↩︎